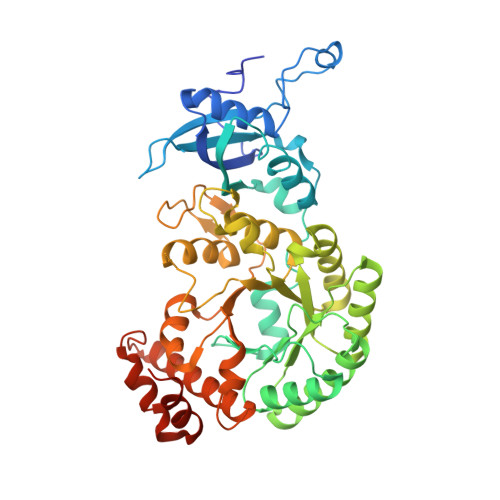
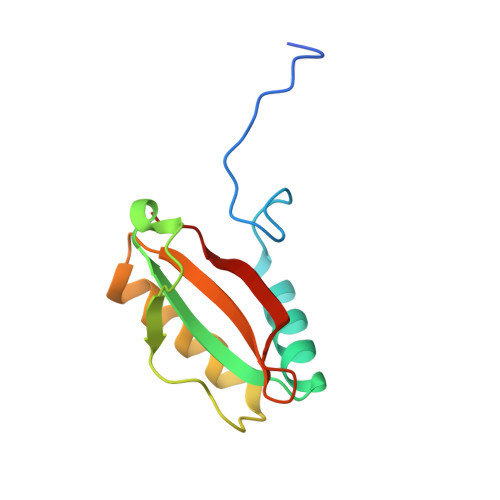
Single-particle cryo-EM analysis of the shell architecture and internal organization of an intact alpha-carboxysome.
Evans, S.L., Al-Hazeem, M.M.J., Mann, D., Smetacek, N., Beavil, A.J., Sun, Y., Chen, T., Dykes, G.F., Liu, L.N., Bergeron, J.R.C.(2023) Structure 31: 677
- PubMed: 37015227
- DOI: https://doi.org/10.1016/j.str.2023.03.008
- Primary Citation of Related Structures:
7YYO, 8CMY - PubMed Abstract:
Carboxysomes are proteinaceous bacterial microcompartments that sequester the key enzymes for carbon fixation in cyanobacteria and some proteobacteria. They consist of a virus-like icosahedral shell, encapsulating several enzymes, including ribulose 1,5-bisphosphate carboxylase/oxygenase (RuBisCO), responsible for the first step of the Calvin-Benson-Bassham cycle. Despite their significance in carbon fixation and great bioengineering potentials, the structural understanding of native carboxysomes is currently limited to low-resolution studies. Here, we report the characterization of a native α-carboxysome from a marine cyanobacterium by single-particle cryoelectron microscopy (cryo-EM). We have determined the structure of its RuBisCO enzyme, and obtained low-resolution maps of its icosahedral shell, and of its concentric interior organization. Using integrative modeling approaches, we have proposed a complete atomic model of an intact carboxysome, providing insight into its organization and assembly. This is critical for a better understanding of the carbon fixation mechanism and toward repurposing carboxysomes in synthetic biology for biotechnological applications.
- Randall Centre for Cell and Molecular Biophysics, King's College London, London, UK.
Organizational Affiliation: