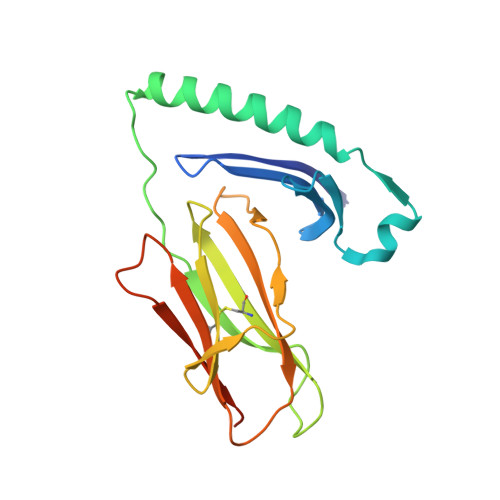
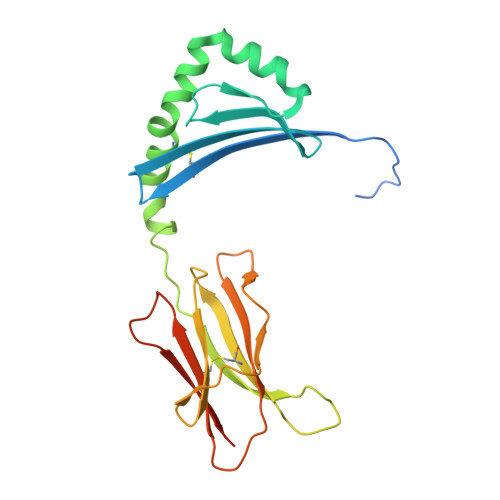
MHC-II dynamics are maintained in HLA-DR allotypes to ensure catalyzed peptide exchange.
Abualrous, E.T., Stolzenberg, S., Sticht, J., Wieczorek, M., Roske, Y., Gunther, M., Dahn, S., Boesen, B.B., Calvo, M.M., Biese, C., Kuppler, F., Medina-Garcia, A., Alvaro-Benito, M., Hofer, T., Noe, F., Freund, C.(2023) Nat Chem Biol 19: 1196-1204
- PubMed: 37142807
- DOI: https://doi.org/10.1038/s41589-023-01316-3
- Primary Citation of Related Structures:
7YX9, 7YXB, 7Z0Q - PubMed Abstract:
Presentation of antigenic peptides by major histocompatibility complex class II (MHC-II) proteins determines T helper cell reactivity. The MHC-II genetic locus displays a large degree of allelic polymorphism influencing the peptide repertoire presented by the resulting MHC-II protein allotypes. During antigen processing, the human leukocyte antigen (HLA) molecule HLA-DM (DM) encounters these distinct allotypes and catalyzes exchange of the placeholder peptide CLIP by exploiting dynamic features of MHC-II. Here, we investigate 12 highly abundant CLIP-bound HLA-DRB1 allotypes and correlate dynamics to catalysis by DM. Despite large differences in thermodynamic stability, peptide exchange rates fall into a target range that maintains DM responsiveness. A DM-susceptible conformation is conserved in MHC-II molecules, and allosteric coupling between polymorphic sites affects dynamic states that influence DM catalysis. As exemplified for rheumatoid arthritis, we postulate that intrinsic dynamic features of peptide-MHC-II complexes contribute to the association of individual MHC-II allotypes with autoimmune disease.
- Protein Biochemistry, Institute for Chemistry and Biochemistry, Freie Universität Berlin, Berlin, Germany.
Organizational Affiliation: