Validation of a New Methodology to Create Oral Drugs beyond the Rule of 5 for Intracellular Tough Targets.
Ohta, A., Tanada, M., Shinohara, S., Morita, Y., Nakano, K., Yamagishi, Y., Takano, R., Kariyuki, S., Iida, T., Matsuo, A., Ozeki, K., Emura, T., Sakurai, Y., Takano, K., Higashida, A., Kojima, M., Muraoka, T., Takeyama, R., Kato, T., Kimura, K., Ogawa, K., Ohara, K., Tanaka, S., Kikuchi, Y., Hisada, N., Hayashi, R., Nishimura, Y., Nomura, K., Tachibana, T., Irie, M., Kawada, H., Torizawa, T., Murao, N., Kotake, T., Tanaka, M., Ishikawa, S., Miyake, T., Tamiya, M., Arai, M., Chiyoda, A., Akai, S., Sase, H., Kuramoto, S., Ito, T., Shiraishi, T., Kojima, T., Iikura, H.(2023) J Am Chem Soc 145: 24035-24051
- PubMed: 37874670
- DOI: https://doi.org/10.1021/jacs.3c07145
- Primary Citation of Related Structures:
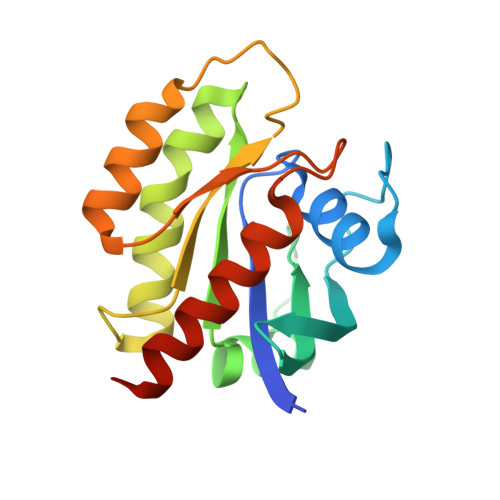
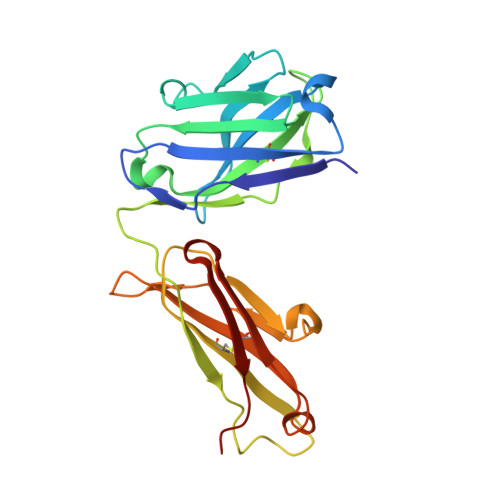
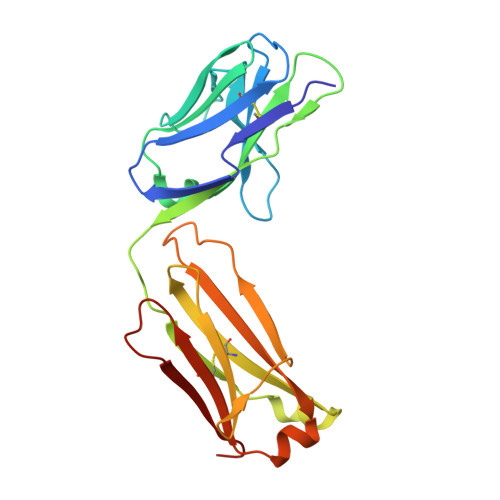
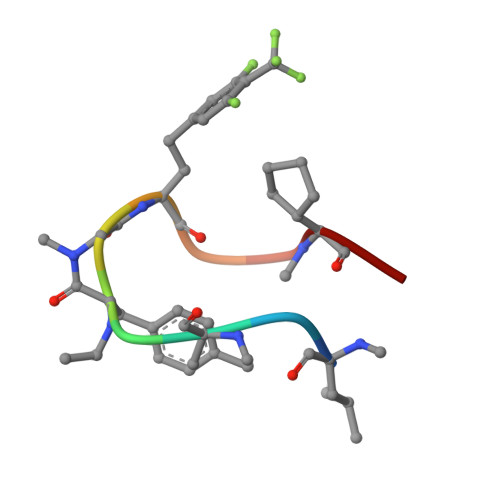
7YUZ, 7YV1 - PubMed Abstract:
Establishing a technological platform for creating clinical compounds inhibiting intracellular protein-protein interactions (PPIs) can open the door to many valuable drugs. Although small molecules and antibodies are mainstream modalities, they are not suitable for a target protein that lacks a deep cavity for a small molecule to bind or a protein found in intracellular space out of an antibody's reach. One possible approach to access these targets is to utilize so-called middle-size cyclic peptides (defined here as those with a molecular weight of 1000-2000 g/mol). In this study, we validated a new methodology to create oral drugs beyond the rule of 5 for intracellular tough targets by elucidating structural features and physicochemical properties for drug-like cyclic peptides and developing library technologies to afford highly N -alkylated cyclic peptide hits. We discovered a KRAS inhibitory clinical compound (LUNA18) as the first example of our platform technology.
- Research Division, Chugai Pharmaceutical Co., Ltd., 216, Totsuka-cho,Totsuka-ku, Yokohama 244-8602, Kanagawa, Japan.
Organizational Affiliation: