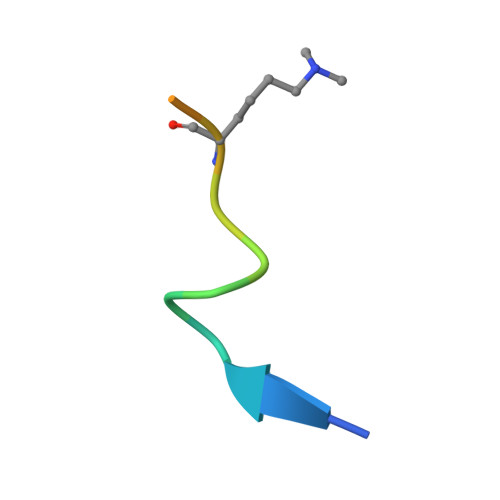
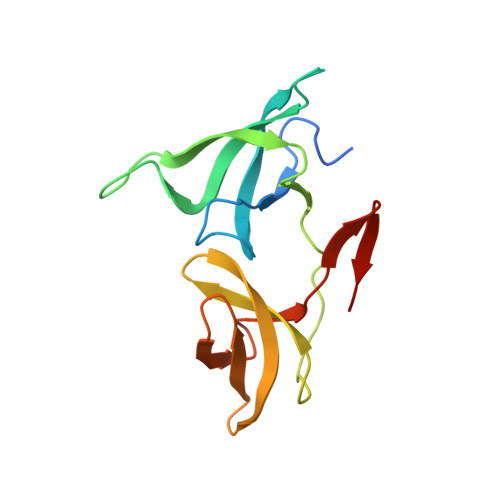The H3K9me2-binding protein AGDP3 limits DNA methylation and transcriptional gene silencing in Arabidopsis.
Zhou, X., Wei, M., Nie, W., Xi, Y., Peng, L., Zheng, Q., Tang, K., Satheesh, V., Wang, Y., Luo, J., Du, X., Liu, R., Yang, Z., La, H., Zhong, Y., Yang, Y., Zhu, J.K., Du, J., Lei, M.(2022) J Integr Plant Biol 64: 2385-2395
- PubMed: 36149781
- DOI: https://doi.org/10.1111/jipb.13369
- Primary Citation of Related Structures:
7YT9, 7YTA - PubMed Abstract:
DNA methylation, a conserved epigenetic mark, is critical for tuning temporal and spatial gene expression. The Arabidopsis thaliana DNA glycosylase/lyase REPRESSOR OF SILENCING 1 (ROS1) initiates active DNA demethylation and is required to prevent DNA hypermethylation at thousands of genomic loci. However, how ROS1 is recruited to specific loci is not well understood. Here, we report the discovery of Arabidopsis AGENET Domain Containing Protein 3 (AGDP3) as a cellular factor that is required to prevent gene silencing and DNA hypermethylation. AGDP3 binds to H3K9me2 marks in its target DNA via its AGD12 cassette. Analysis of the crystal structure of the AGD12 cassette of AGDP3 in complex with an H3K9me2 peptide revealed that dimethylated H3K9 and unmodified H3K4 are specifically anchored into two different surface pockets. A histidine residue located in the methyllysine binding aromatic cage provides AGDP3 with pH-dependent H3K9me2 binding capacity. Our results uncover a molecular mechanism for the regulation of DNA demethylation by the gene silencing mark H3K9me2.
- Shanghai Center for Plant Stress Biology, CAS Center for Excellence in Molecular Plant Sciences, Chinese Academy of Sciences, Shanghai, 201602, China.
Organizational Affiliation: