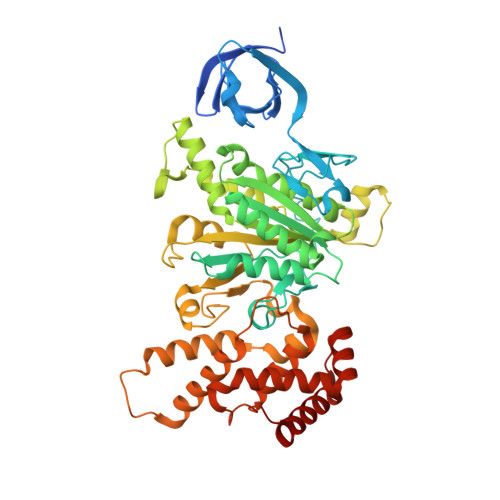
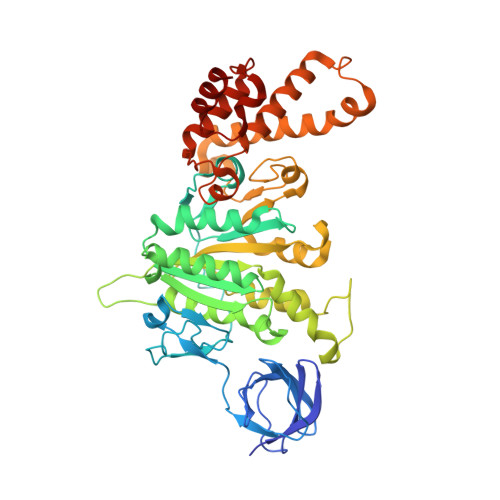
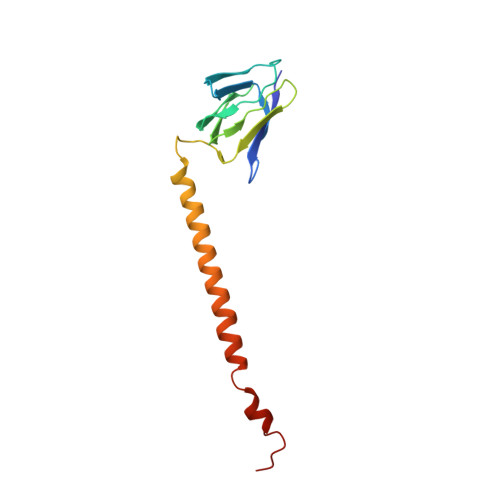
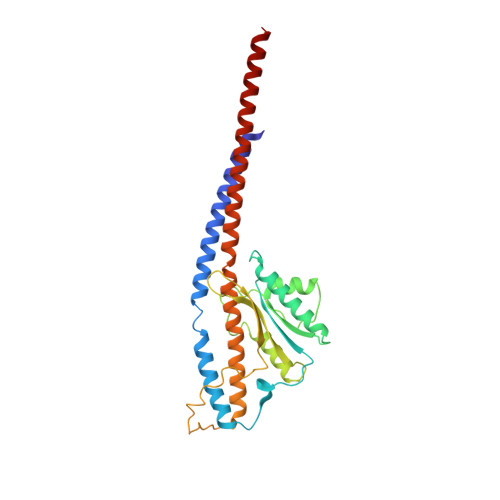
Atomic insights of an up and down conformation of the Acinetobacter baumannii F 1 -ATPase subunit epsilon and deciphering the residues critical for ATP hydrolysis inhibition and ATP synthesis.
Saw, W.G., Le, K.C.M., Shin, J., Kwek, J.H.M., Wong, C.F., Ragunathan, P., Fong, T.C., Muller, V., Gruber, G.(2023) FASEB J 37: e23040-e23040
- PubMed: 37318822
- DOI: https://doi.org/10.1096/fj.202300175RR
- Primary Citation of Related Structures:
7YRY, 8HGX - PubMed Abstract:
The Acinetobacter baumannii F 1 F O -ATP synthase (α 3 :β 3 :γ:δ:ε:a:b 2 :c 10 ), which is essential for this strictly respiratory opportunistic human pathogen, is incapable of ATP-driven proton translocation due to its latent ATPase activity. Here, we generated and purified the first recombinant A. baumannii F 1 -ATPase (AbF 1 -ATPase) composed of subunits α 3 :β 3 :γ:ε, showing latent ATP hydrolysis. A 3.0 Å cryo-electron microscopy structure visualizes the architecture and regulatory element of this enzyme, in which the C-terminal domain of subunit ε (Abε) is present in an extended position. An ε-free AbF 1 -ɑβγ complex generated showed a 21.5-fold ATP hydrolysis increase, demonstrating that Abε is the major regulator of AbF 1 -ATPase's latent ATP hydrolysis. The recombinant system enabled mutational studies of single amino acid substitutions within Abε or its interacting subunits β and γ, respectively, as well as C-terminal truncated mutants of Abε, providing a detailed picture of Abε's main element for the self-inhibition mechanism of ATP hydrolysis. Using a heterologous expression system, the importance of Abε's C-terminus in ATP synthesis of inverted membrane vesicles, including AbF 1 F O -ATP synthases, has been explored. In addition, we are presenting the first NMR solution structure of the compact form of Abε, revealing interaction of its N-terminal β-barrel and C-terminal ɑ-hairpin domain. A double mutant of Abε highlights critical residues for Abε's domain-domain formation which is important also for AbF 1 -ATPase's stability. Abε does not bind MgATP, which is described to regulate the up and down movements in other bacterial counterparts. The data are compared to regulatory elements of F 1 -ATPases in bacteria, chloroplasts, and mitochondria to prevent wasting of ATP.
- School of Biological Sciences, Nanyang Technological University, Singapore, Singapore.
Organizational Affiliation: