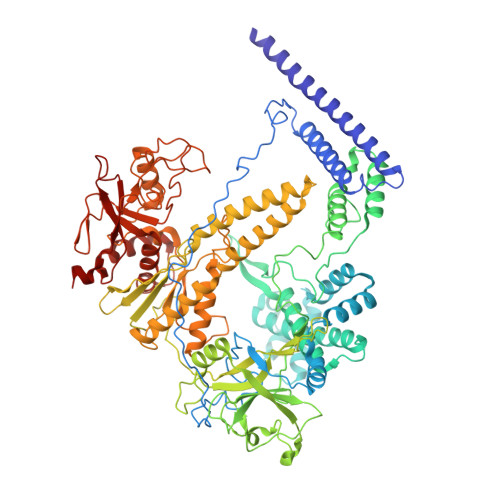
The compact Cas pi (Cas12l) 'bracelet' provides a unique structural platform for DNA manipulation.
Sun, A., Li, C.P., Chen, Z., Zhang, S., Li, D.Y., Yang, Y., Li, L.Q., Zhao, Y., Wang, K., Li, Z., Liu, J., Liu, S., Wang, J., Liu, J.G.(2023) Cell Res 33: 229-244
- PubMed: 36650285
- DOI: https://doi.org/10.1038/s41422-022-00771-2
- Primary Citation of Related Structures:
7YOJ - PubMed Abstract:
CRISPR-Cas modules serve as the adaptive nucleic acid immune systems for prokaryotes, and provide versatile tools for nucleic acid manipulation in various organisms. Here, we discovered a new miniature type V system, CRISPR-Casπ (Cas12l) (~860 aa), from the environmental metagenome. Complexed with a large guide RNA (~170 nt) comprising the tracrRNA and crRNA, Casπ (Cas12l) recognizes a unique 5' C-rich PAM for DNA cleavage under a broad range of biochemical conditions, and generates gene editing in mammalian cells. Cryo-EM study reveals a 'bracelet' architecture of Casπ effector encircling the DNA target at 3.4 Å resolution, substantially different from the canonical 'two-lobe' architectures of Cas12 and Cas9 nucleases. The large guide RNA serves as a 'two-arm' scaffold for effector assembly. Our study expands the knowledge of DNA targeting mechanisms by CRISPR effectors, and offers an efficient but compact platform for DNA manipulation.
- Beijing Advanced Innovation Center for Structural Biology & Frontier Research Center for Biological Structure, School of Life Sciences, Tsinghua University, Beijing, China.
Organizational Affiliation: