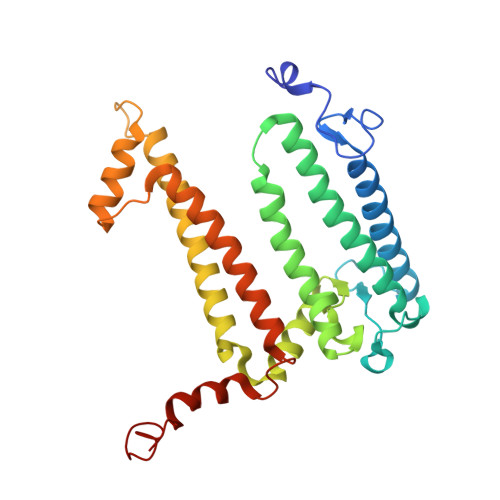
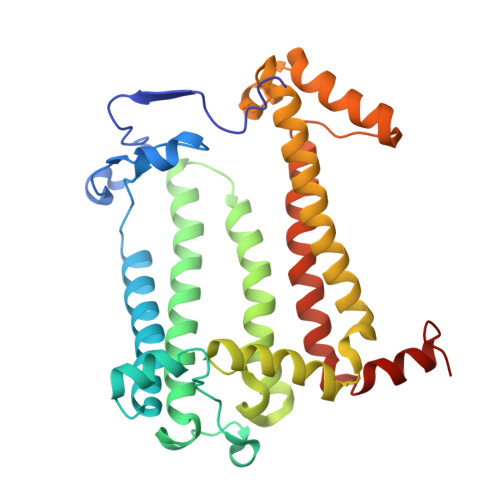
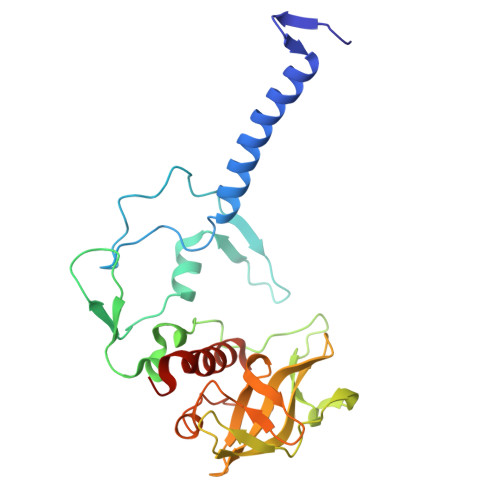
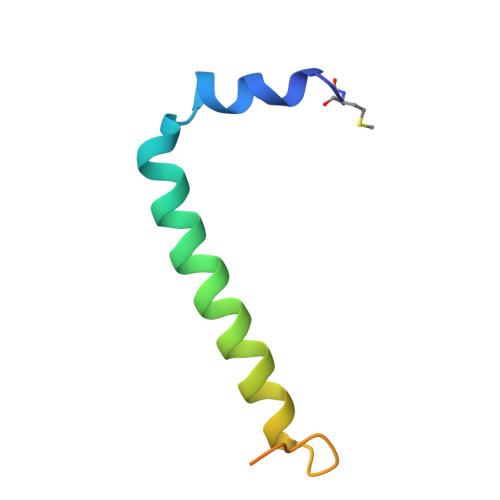
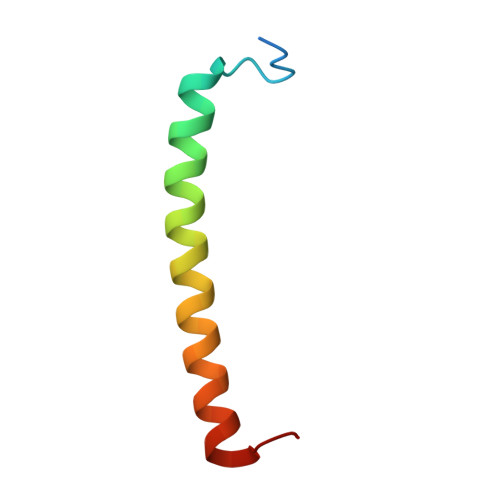
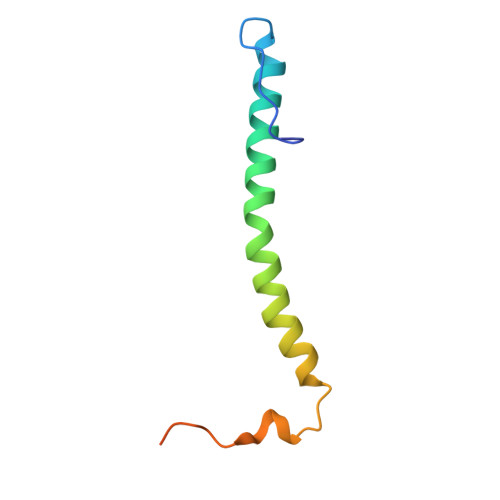
Rhodobacter capsulatus forms a compact crescent-shaped LH1-RC photocomplex.
Tani, K., Kanno, R., Ji, X.C., Satoh, I., Kobayashi, Y., Hall, M., Yu, L.J., Kimura, Y., Mizoguchi, A., Humbel, B.M., Madigan, M.T., Wang-Otomo, Z.Y.(2023) Nat Commun 14: 846-846
- PubMed: 36792596
- DOI: https://doi.org/10.1038/s41467-023-36460-w
- Primary Citation of Related Structures:
7YML - PubMed Abstract:
Rhodobacter (Rba.) capsulatus has been a favored model for studies of all aspects of bacterial photosynthesis. This purple phototroph contains PufX, a polypeptide crucial for dimerization of the light-harvesting 1-reaction center (LH1-RC) complex, but lacks protein-U, a U-shaped polypeptide in the LH1-RC of its close relative Rba. sphaeroides. Here we present a cryo-EM structure of the Rba. capsulatus LH1-RC purified by DEAE chromatography. The crescent-shaped LH1-RC exhibits a compact structure containing only 10 LH1 αβ-subunits. Four αβ-subunits corresponding to those adjacent to protein-U in Rba. sphaeroides were absent. PufX in Rba. capsulatus exhibits a unique conformation in its N-terminus that self-associates with amino acids in its own transmembrane domain and interacts with nearby polypeptides, preventing it from interacting with proteins in other complexes and forming dimeric structures. These features are discussed in relation to the minimal requirements for the formation of LH1-RC monomers and dimers, the spectroscopic behavior of both the LH1 and RC, and the bioenergetics of energy transfer from LH1 to the RC.
- Graduate School of Medicine, Mie University, Tsu, Japan. ktani@doc.medic.mie-u.ac.jp.
Organizational Affiliation: