Structural and Functional Analysis of the Pyridoxal Phosphate Homeostasis Protein YggS from Fusobacterium nucleatum.
He, S., Chen, Y., Wang, L., Bai, X., Bu, T., Zhang, J., Lu, M., Ha, N.C., Quan, C., Nam, K.H., Xu, Y.(2022) Molecules 27
- PubMed: 35897955
- DOI: https://doi.org/10.3390/molecules27154781
- Primary Citation of Related Structures:
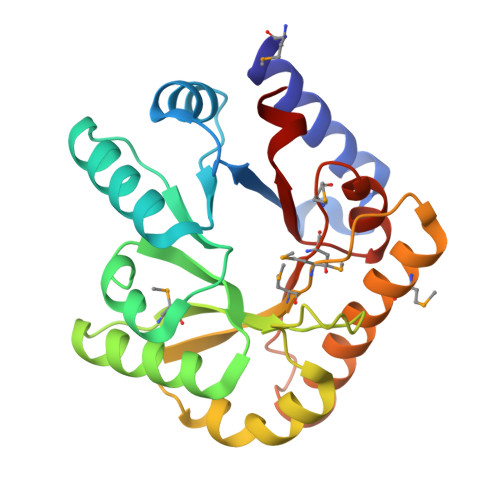
7YGF - PubMed Abstract:
Pyridoxal 5′-phosphate (PLP) is the active form of vitamin B6, but it is highly reactive and poisonous in its free form. YggS is a PLP-binding protein found in bacteria and humans that mediates PLP homeostasis by delivering PLP to target enzymes or by performing a protective function. Several biochemical and structural studies of YggS have been reported, but the mechanism by which YggS recognizes PLP has not been fully elucidated. Here, we report a functional and structural analysis of YggS from Fusobacterium nucleatum (FnYggS). The PLP molecule could bind to native FnYggS, but no PLP binding was observed for selenomethionine (SeMet)-derivatized FnYggS. The crystal structure of FnYggS showed a type III TIM barrel fold, exhibiting structural homology with several other PLP-dependent enzymes. Although FnYggS exhibited low (<35%) amino acid sequence similarity with previously studied YggS proteins, its overall structure and PLP-binding site were highly conserved. In the PLP-binding site of FnYggS, the sulfate ion was coordinated by the conserved residues Ser201, Gly218, and Thr219, which were positioned to provide the binding moiety for the phosphate group of PLP. The mutagenesis study showed that the conserved Ser201 residue in FnYggS was the key residue for PLP binding. These results will expand the knowledge of the molecular properties and function of the YggS family.
- Department of Bioengineering, College of Life Science, Dalian Minzu University, Dalian 116600, China.
Organizational Affiliation: