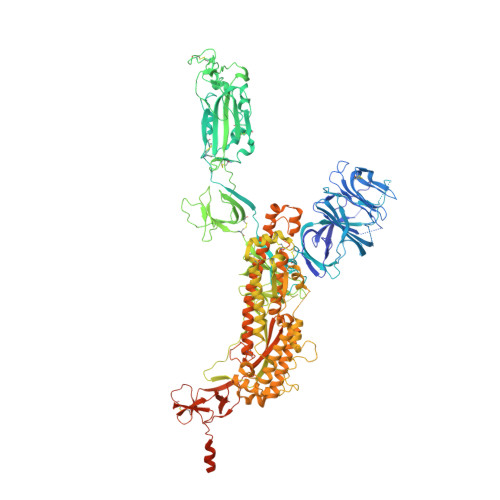
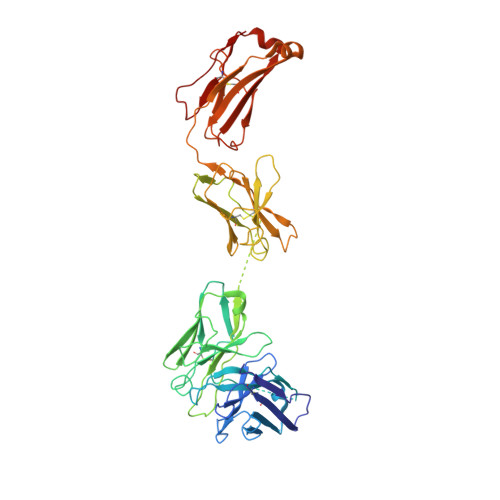
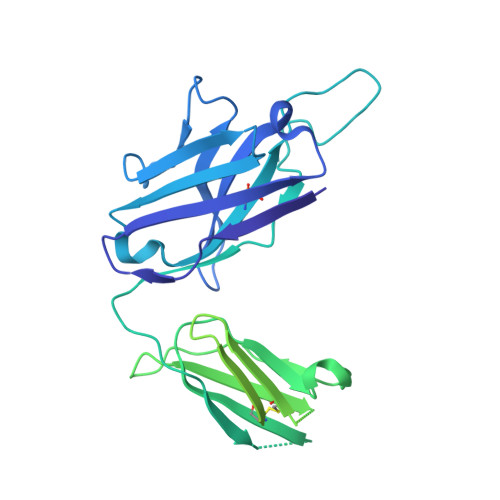
Novel bispecific human antibody platform specifically targeting a fully open spike conformation potently neutralizes multiple SARS-CoV-2 variants
Kim, J.W., Heo, K., Kim, H.J., Yoo, Y., Cho, H.S., Jang, H.J., Lee, H.Y., Ko, I.Y., Woo, J.R., Cho, Y.B., Lee, J.H., Yang, H.R., Shin, H.G., Choi, H.L., Hwang, K., Kim, S., Kim, H., Chun, K., Lee, S.(2023) Antiviral Res 212: 105576