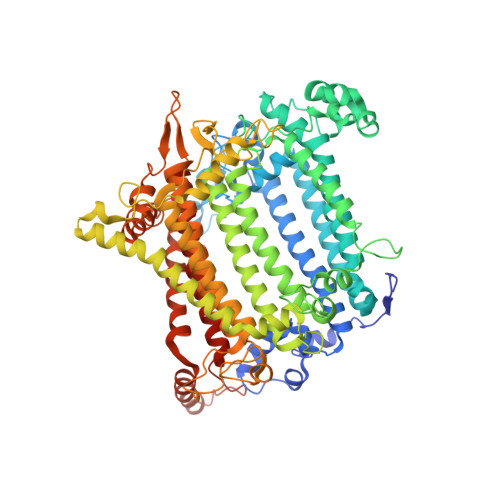
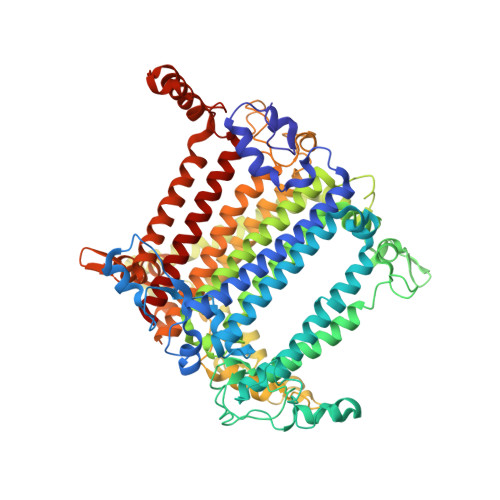
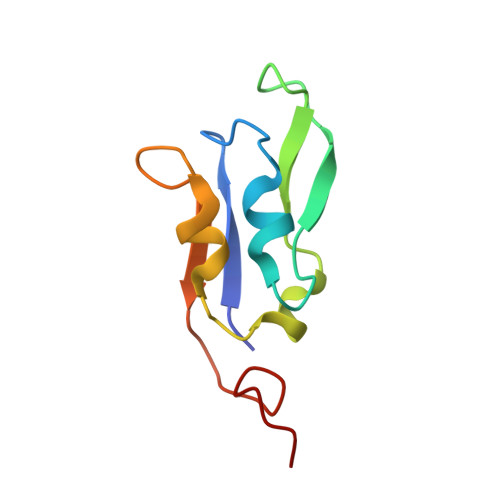
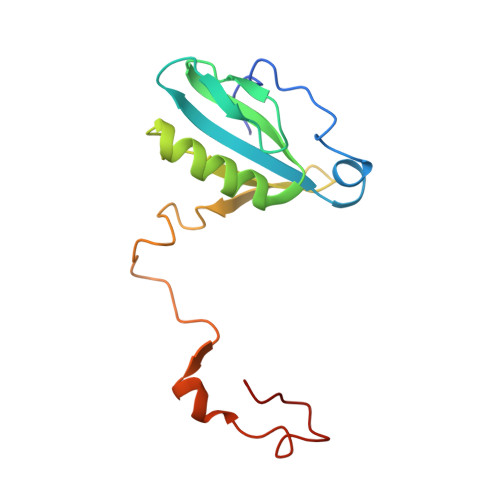
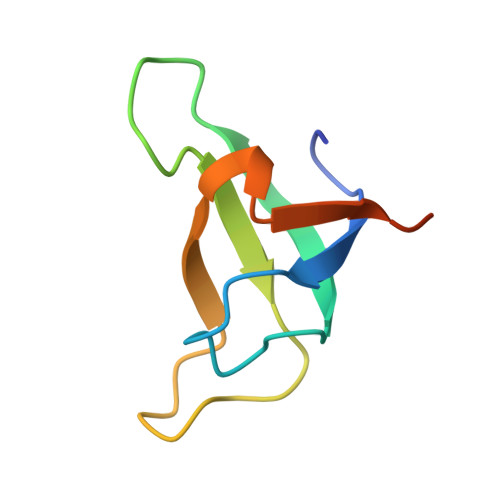
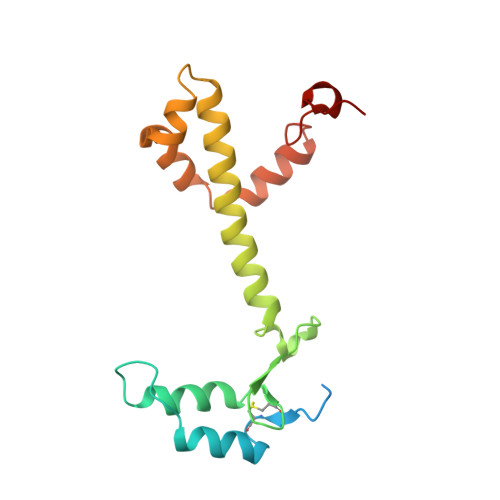
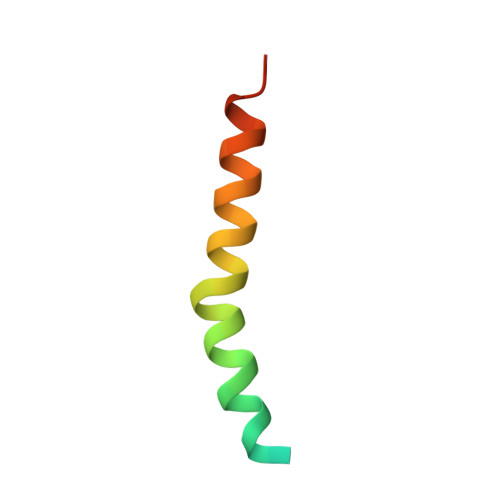
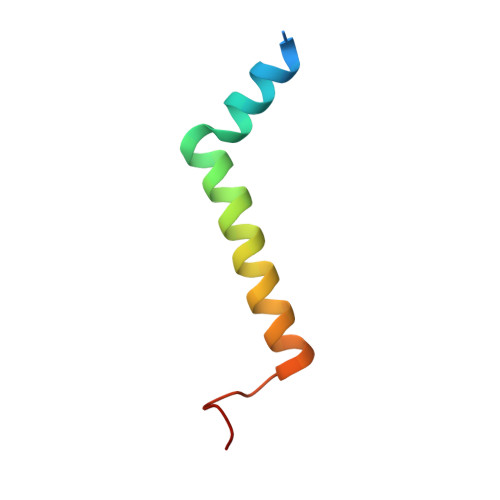
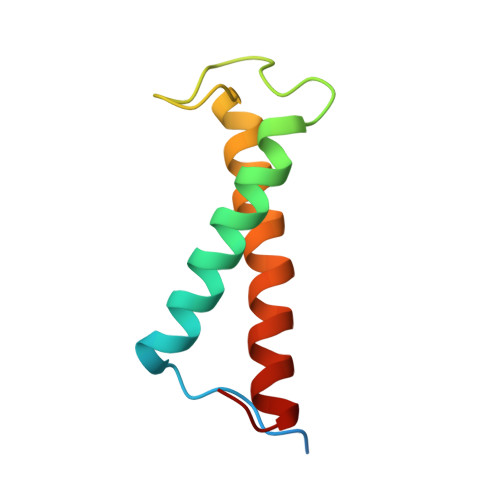
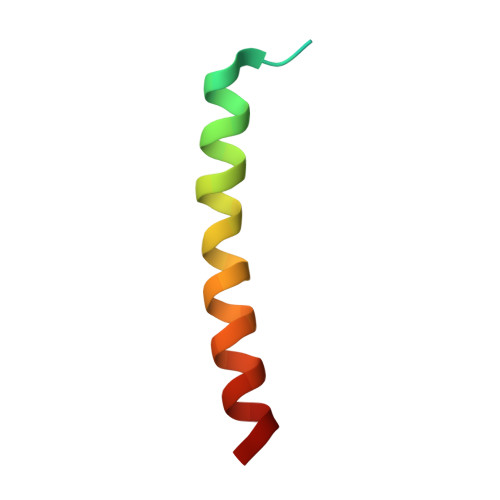
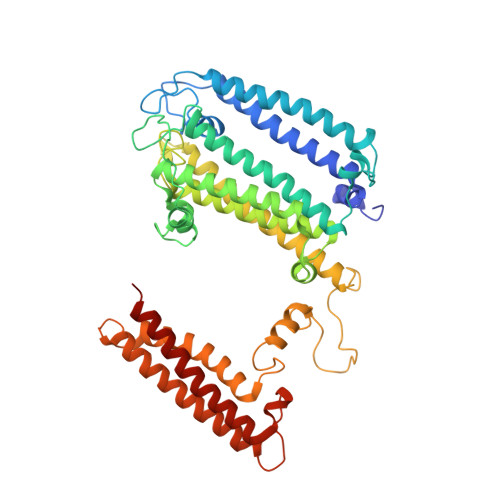
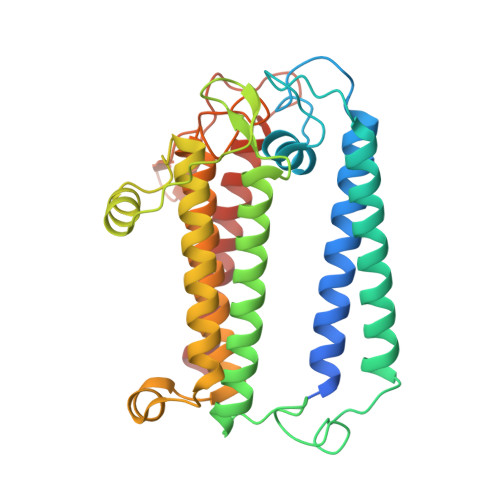
Structure of a monomeric photosystem I core associated with iron-stress-induced-A proteins from Anabaena sp. PCC 7120.
Nagao, R., Kato, K., Hamaguchi, T., Ueno, Y., Tsuboshita, N., Shimizu, S., Furutani, M., Ehira, S., Nakajima, Y., Kawakami, K., Suzuki, T., Dohmae, N., Akimoto, S., Yonekura, K., Shen, J.R.(2023) Nat Commun 14: 920-920
- PubMed: 36805598
- DOI: https://doi.org/10.1038/s41467-023-36504-1
- Primary Citation of Related Structures:
7Y3F - PubMed Abstract:
Iron-stress-induced-A proteins (IsiAs) are expressed in cyanobacteria under iron-deficient conditions. The cyanobacterium Anabaena sp. PCC 7120 has four isiA genes; however, their binding property and functional roles in PSI are still missing. We analyzed a cryo-electron microscopy structure of a PSI-IsiA supercomplex isolated from Anabaena grown under an iron-deficient condition. The PSI-IsiA structure contains six IsiA subunits associated with the PsaA side of a PSI core monomer. Three of the six IsiA subunits were identified as IsiA1 and IsiA2. The PSI-IsiA structure lacks a PsaL subunit; instead, a C-terminal domain of IsiA2 occupies the position of PsaL, which inhibits the oligomerization of PSI, leading to the formation of a PSI monomer. Furthermore, excitation-energy transfer from IsiAs to PSI appeared with a time constant of 55 ps. These findings provide insights into both the molecular assembly of the Anabaena IsiA family and the functional roles of IsiAs.
- Research Institute for Interdisciplinary Science and Graduate School of Natural Science and Technology, Okayama University, Okayama, 700-8530, Japan. nagao.ryo@shizuoka.ac.jp.
Organizational Affiliation: