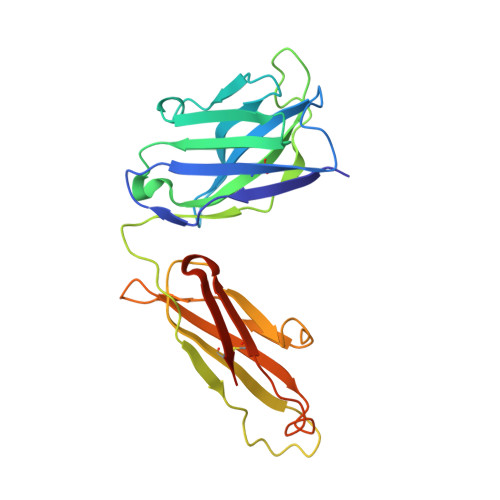
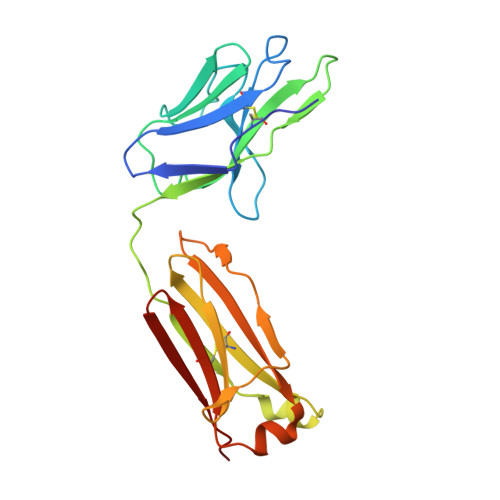
Rational identification of potent and broad sarbecovirus-neutralizing antibody cocktails from SARS convalescents.
Cao, Y., Jian, F., Zhang, Z., Yisimayi, A., Hao, X., Bao, L., Yuan, F., Yu, Y., Du, S., Wang, J., Xiao, T., Song, W., Zhang, Y., Liu, P., An, R., Wang, P., Wang, Y., Yang, S., Niu, X., Zhang, Y., Gu, Q., Shao, F., Hu, Y., Yin, W., Zheng, A., Wang, Y., Qin, C., Jin, R., Xiao, J., Xie, X.S.(2022) Cell Rep 41: 111845-111845
- PubMed: 36493787
- DOI: https://doi.org/10.1016/j.celrep.2022.111845
- Primary Citation of Related Structures:
7WRJ, 7WRY, 7Y0C, 7Y0V, 7Y0W - PubMed Abstract:
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Omicron sublineages have escaped most receptor-binding domain (RBD)-targeting therapeutic neutralizing antibodies (NAbs), which proves that previous NAb drug screening strategies are deficient against the fast-evolving SARS-CoV-2. Better broad NAb drug candidate selection methods are needed. Here, we describe a rational approach for identifying RBD-targeting broad SARS-CoV-2 NAb cocktails. Based on high-throughput epitope determination, we propose that broad NAb drugs should target non-immunodominant RBD epitopes to avoid herd-immunity-directed escape mutations. Also, their interacting antigen residues should focus on sarbecovirus conserved sites and associate with critical viral functions, making the antibody-escaping mutations less likely to appear. Following these criteria, a featured non-competing antibody cocktail, SA55+SA58, is identified from a large collection of broad sarbecovirus NAbs isolated from SARS-CoV-2-vaccinated SARS convalescents. SA55+SA58 potently neutralizes ACE2-utilizing sarbecoviruses, including circulating Omicron variants, and could serve as broad SARS-CoV-2 prophylactics to offer long-term protection, especially for individuals who are immunocompromised or with high-risk comorbidities.
- Biomedical Pioneering Innovation Center (BIOPIC), Peking University, Beijing, P.R. China; Changping Laboratory, Beijing, P.R. China. Electronic address: yunlongcao@pku.edu.cn.
Organizational Affiliation: