Extension Peptide of Plant Ferritin from Setaria italica Presents a Novel Fold.
Wang, W., Wang, Y., Xi, H., Song, Z., Zhang, W., Xie, L., Ma, D., Qin, N., Wang, H.(2023) J Agric Food Chem 71: 934-943
- PubMed: 36576327
- DOI: https://doi.org/10.1021/acs.jafc.2c07595
- Primary Citation of Related Structures:
7XZ4 - PubMed Abstract:
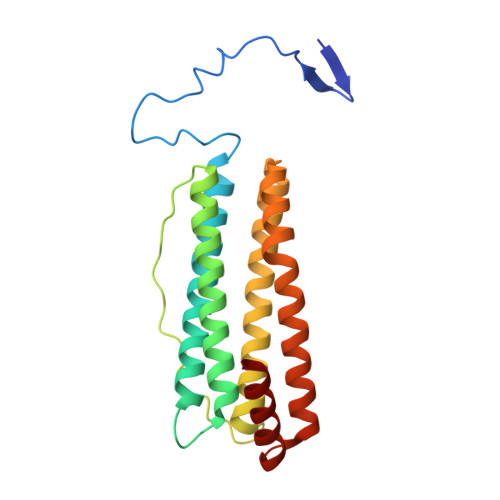
The extension peptide (EP) is the most distinctive feature of mature plant ferritin. Some EPs have exhibited serine-like protease activity, which is associated with iron uptake and release. EP forms a helix and a long loop, followed by a conserved core helical bundle. However, whether the EP adopts a stable or uniform folding pattern in all plants remains unclear. To clarify this, we investigated the crystal structure of ferritin-1 from Setaria italica (SiFer1), a type of monocotyledon. In our structure of SiFer1, the EP is different from other EPs in other solved structures of plant ferritins and consisted of a pair of β-sheets, a shorter helix, and two loops, which masks two hydrophobic pockets on the outer surface of every subunit. Furthermore, sequence analysis and structure comparison suggest that the EPs in ferritins from monocotyledons may adopt a novel fold pattern, and the conformations of EPs in ferritins are alterable among different plant species. Furthermore, additional eight iron atoms were first founded binding in the fourfold channels, demonstrating the vital function of fourfold channels in iron diffusion. In all, our structure provides new clues for understanding plant ferritins and the functions of the EP.
Organizational Affiliation:
Institute of Molecular Science, Shanxi University, Taiyuan 030006, China.