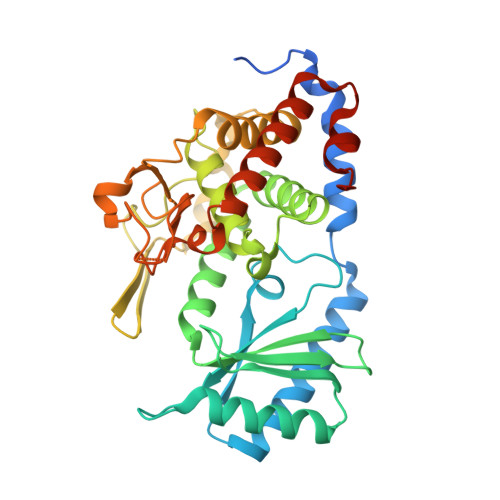
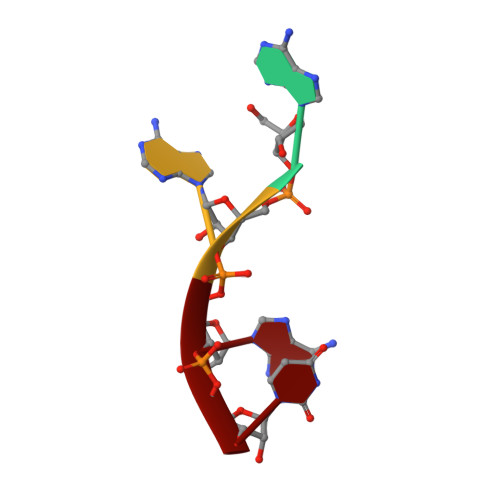
Molecular mechanism underlying the di-uridylation activity of Arabidopsis TUTase URT1.
Hu, Q., Yang, H., Li, M., Zhu, L., Lv, M., Li, F., Zhang, Z., Ren, G., Gong, Q.(2022) Nucleic Acids Res 50: 10614-10625
- PubMed: 36177876
- DOI: https://doi.org/10.1093/nar/gkac839
- Primary Citation of Related Structures:
7XS4 - PubMed Abstract:
In Arabidopsis, HESO1 and URT1 act cooperatively on unmethylated miRNA and mRNA uridylation to induce their degradation. Their collaboration significantly impacts RNA metabolism in plants. However, the molecular mechanism determining the functional difference and complementarity of these two enzymes remains unclear. We previously solved the three-dimensional structure of URT1 in the absence and presence of UTP. In this study, we further determined the structure of URT1 in complex with a 5'-AAAU-3' RNA stretch that mimics the post-catalytic state of the mRNA poly(A) tail after the addition of the first uridine. Structural analysis and enzymatic assays revealed that L527 and Y592 endow URT1 with a preference to interact with purine over pyrimidine at the -1 RNA binding position, thus controlling the optimal number of uridine added to the 3' extremity of poly(A) as two. In addition, we observed that a large-scale conformational rearrangement in URT1 occurs upon binding with RNA from an 'open' to a 'closed' state. Molecular dynamic simulation supports an open-closed conformational selection mechanism employed by URT1 to interact with RNA substrates and maintain distributive enzymatic activity. Based on the above results, a model regarding the catalytic cycle of URT1 is proposed to explain its di-uridylation activity.
- Department of Clinical Laboratory, The First Affiliated Hospital of USTC, Ministry of Education Key Laboratory for Membraneless Organelles & Cellular Dynamics, Biomedical Sciences and Health Laboratory of Anhui Province, School of Life Sciences, Division of Life Sciences and Medicine, University of Science and Technology of China, 230027 Hefei, P.R. China.
Organizational Affiliation: