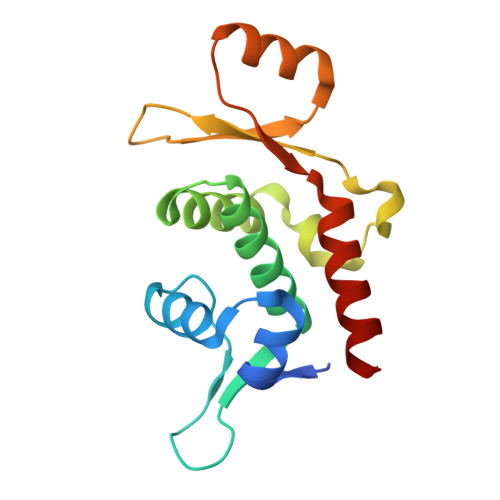
CvkR is a MerR-type transcriptional repressor of class 2 type V-K CRISPR-associated transposase systems.
Ziemann, M., Reimann, V., Liang, Y., Shi, Y., Ma, H., Xie, Y., Li, H., Zhu, T., Lu, X., Hess, W.R.(2023) Nat Commun 14: 924-924
- PubMed: 36801863
- DOI: https://doi.org/10.1038/s41467-023-36542-9
- Primary Citation of Related Structures:
7XN2 - PubMed Abstract:
Certain CRISPR-Cas elements integrate into Tn7-like transposons, forming CRISPR-associated transposon (CAST) systems. How the activity of these systems is controlled in situ has remained largely unknown. Here we characterize the MerR-type transcriptional regulator Alr3614 that is encoded by one of the CAST (AnCAST) system genes in the genome of cyanobacterium Anabaena sp. PCC 7120. We identify a number of Alr3614 homologs across cyanobacteria and suggest naming these regulators CvkR for Cas V-K repressors. Alr3614/CvkR is translated from leaderless mRNA and represses the AnCAST core modules cas12k and tnsB directly, and indirectly the abundance of the tracr-CRISPR RNA. We identify a widely conserved CvkR binding motif 5'-AnnACATnATGTnnT-3'. Crystal structure of CvkR at 1.6 Å resolution reveals that it comprises distinct dimerization and potential effector-binding domains and that it assembles into a homodimer, representing a discrete structural subfamily of MerR regulators. CvkR repressors are at the core of a widely conserved regulatory mechanism that controls type V-K CAST systems.
- Faculty of Biology, Institute of Biology III, Genetics and Experimental Bioinformatics, University of Freiburg, Schänzlestr. 1, Freiburg, D-79104, Germany.
Organizational Affiliation: