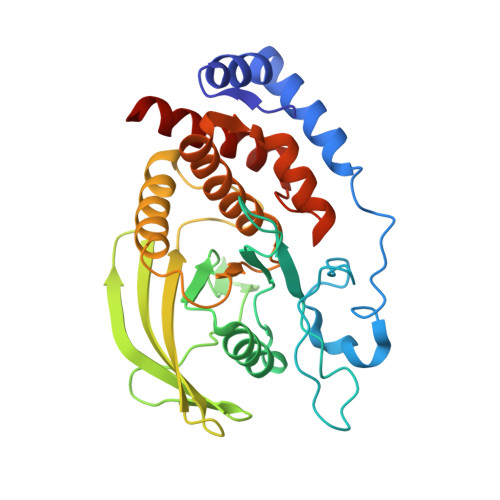
Crystal structure of the catalytic domain of human RPTPH.
Kim, M., Ryu, S.E.(2022) Acta Crystallogr F Struct Biol Commun 78: 265-269
- PubMed: 35787553
- DOI: https://doi.org/10.1107/S2053230X22006173
- Primary Citation of Related Structures:
7XC0 - PubMed Abstract:
Receptor-type protein tyrosine phosphatases (RPTPs) receive extracellular stimuli and transfer them into cells. They regulate cell growth, differentiation and death via specific signals. They have also been implicated in cancer, diabetes and neurological diseases. RPTPH, a member of the type 3 RPTP (R3-PTP) family, is an important regulator of colorectal cancer and hepatic carcinoma. Despite its importance in drug development, the structure of RPTPH has not yet been resolved. Here, the crystal structure of the catalytic domain of RPTPH was determined at 1.56 Å resolution. Despite similarities to other R3-PTPs in its overall structure, RPTPH exhibited differences in its loop regions and side-chain conformations. Compared with other R3-PTPs, RPTPH has unique side chains near its active site that may confer specificity for inhibitor binding. Therefore, detailed information on the structure of RPTPH provides clues for the development of specific inhibitors.
- Department of Bioengineering, College of Engineering, Hanyang University, Seoul 04673, Republic of Korea.
Organizational Affiliation: