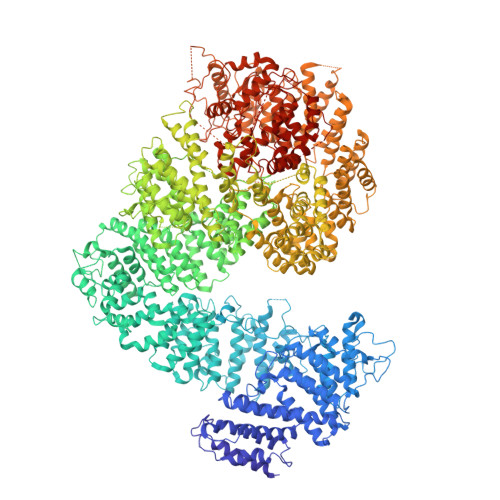
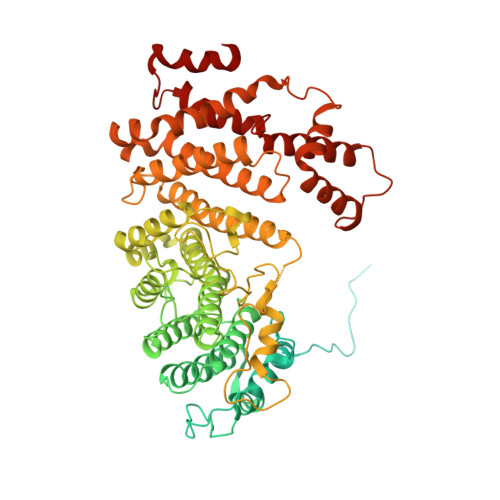
Structures of Mec1/ATR kinase endogenously stimulated by different genotoxins.
Zhang, Q., Wang, P., Wu, T., Zhang, Y., Zheng, Z., Zhou, S., Qian, D., Wang, X., Cai, G.(2022) Cell Discov 8: 98-98
- PubMed: 36175395
- DOI: https://doi.org/10.1038/s41421-022-00461-8
- Primary Citation of Related Structures:
7WZR, 7WZW - The First Affiliated Hospital of USTC, MOE Key Laboratory for Cellular Dynamics, Division of Life Sciences and Medicine, University of Science and Technology of China, Hefei, Anhui, China.
Organizational Affiliation: