Molecular insights into biogenesis of glycosylphosphatidylinositol anchor proteins.
Xu, Y., Jia, G., Li, T., Zhou, Z., Luo, Y., Chao, Y., Bao, J., Su, Z., Qu, Q., Li, D.(2022) Nat Commun 13: 2617-2617
- PubMed: 35551457
- DOI: https://doi.org/10.1038/s41467-022-30250-6
- Primary Citation of Related Structures:
7WLD - PubMed Abstract:
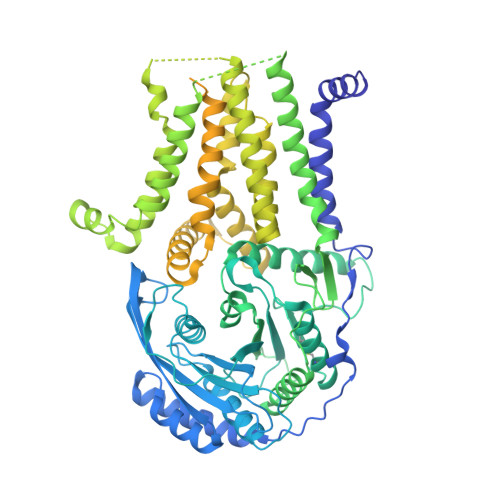
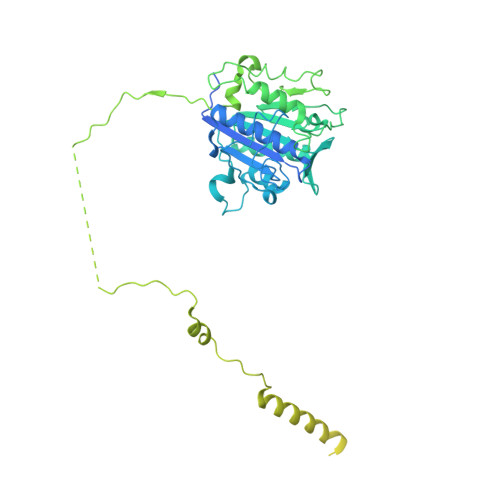
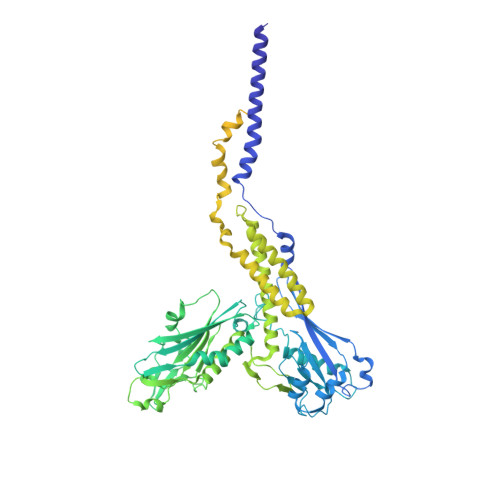
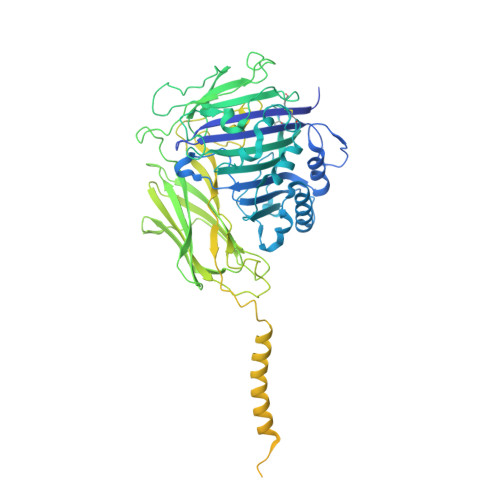
Eukaryotic cells are coated with an abundance of glycosylphosphatidylinositol anchor proteins (GPI-APs) that play crucial roles in fertilization, neurogenesis, and immunity. The removal of a hydrophobic signal peptide and covalent attachment of GPI at the new carboxyl terminus are catalyzed by an endoplasmic reticulum membrane GPI transamidase complex (GPI-T) conserved among all eukaryotes. Here, we report the cryo-electron microscopy (cryo-EM) structure of the human GPI-T at a global 2.53-Å resolution, revealing an equimolar heteropentameric assembly. Structure-based mutagenesis suggests a legumain-like mechanism for the recognition and cleavage of proprotein substrates, and an endogenous GPI in the structure defines a composite cavity for the lipid substrate. This elongated active site, stemming from the membrane and spanning an additional ~22-Å space toward the catalytic dyad, is structurally suited for both substrates which feature an amphipathic pattern that matches this geometry. Our work presents an important step towards the mechanistic understanding of GPI-AP biosynthesis.
- State Key Laboratory of Molecular Biology, CAS Center for Excellence in Molecular Cell Science, Shanghai Institute of Biochemistry and Cell Biology, University of CAS, Chinese Academy of Sciences (CAS), 320 Yueyang Road, 200030, Shanghai, China.
Organizational Affiliation: