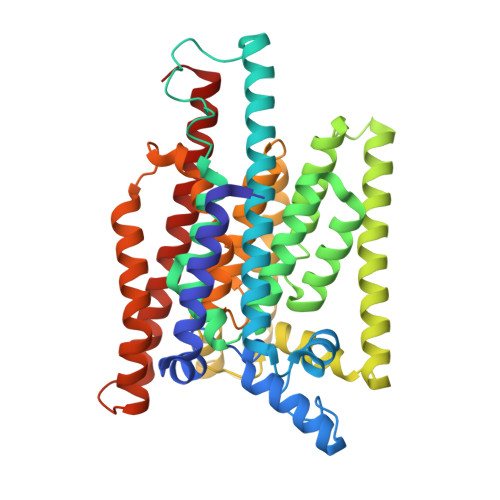
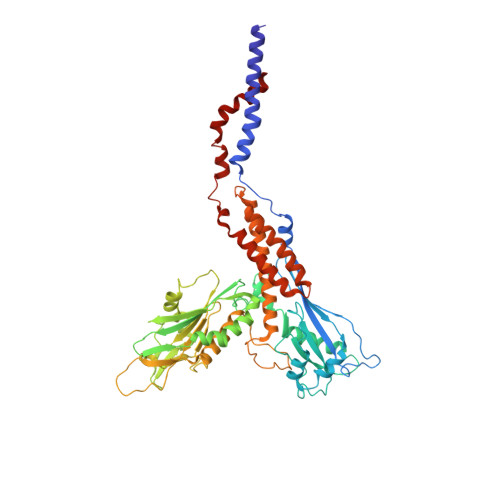
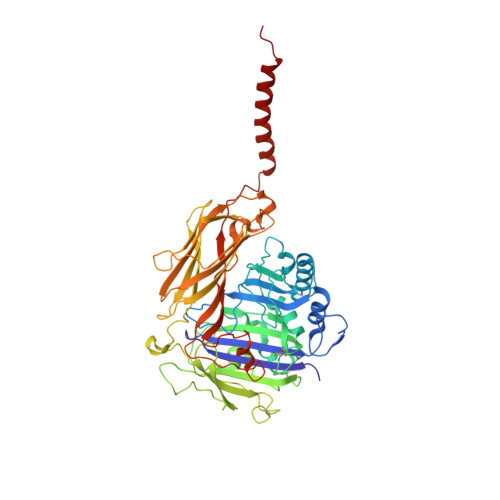
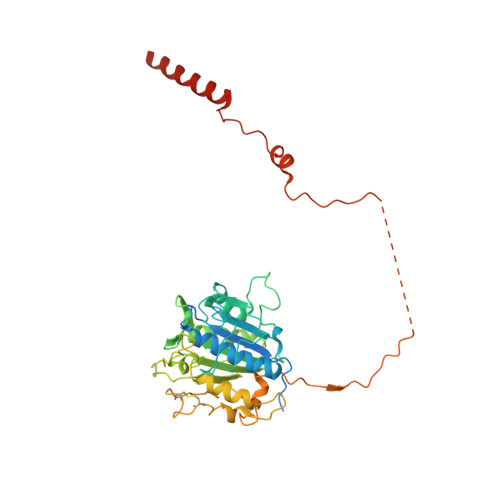
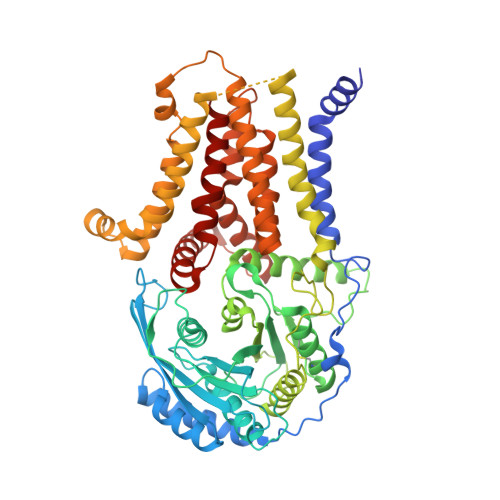
Structure of human glycosylphosphatidylinositol transamidase.
Zhang, H., Su, J., Li, B., Gao, Y., Liu, M., He, L., Xu, H., Dong, Y., Zhang, X.C., Zhao, Y.(2022) Nat Struct Mol Biol 29: 203-209
- PubMed: 35165458
- DOI: https://doi.org/10.1038/s41594-022-00726-6
- Primary Citation of Related Structures:
7W72 - PubMed Abstract:
Glycosylphosphatidylinositol (GPI) molecules are complex glycophospholipids and serve as membrane anchors for tethering many proteins to the cell surface. Attaching GPI to the protein in the endoplasmic reticulum (ER) is catalyzed by the transmembrane GPI transamidase (GPIT) complex, which is essential for maturation of the GPI-anchored proteins. The GPIT complex is known to be composed of five subunits: PIGK, PIGU, PIGT, PIGS and GPAA1. Here, we determined the structure of the human GPIT complex at a resolution of 3.1 Å using single-particle cryo-EM, elucidating its overall assembly. The PIGK subunit functions as the catalytic component, in which we identified a C206-H164-N58 triad that is critical for the transamination reaction. Transmembrane helices constitute a widely opened cleft, which is located underneath PIGK, serving as a GPI substrate-binding site. The ubiquitin E3 ligase RNF121 is visualized at the back of the complex and probably serves as a quality control factor for the GPIT complex.
- National Laboratory of Biomacromolecules, CAS Center for Excellence in Biomacromolecules, Institute of Biophysics, Chinese Academy of Sciences, Beijing, China.
Organizational Affiliation: