Binding and structural basis of equine ACE2 to RBDs from SARS-CoV, SARS-CoV-2 and related coronaviruses
Xu, Z., Kang, X., Han, P., Du, P., Li, L., Zheng, A., Deng, C., Qi, J., Zhao, X., Wang, Q., Liu, K., Gao, G.F.(2022) Nat Commun 13: 3547
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Angiotensin-converting enzyme | 602 | Equus caballus | Mutation(s): 0 Gene Names: ACE2 EC: 3.4.17.23 (PDB Primary Data), 3.4 (UniProt) | 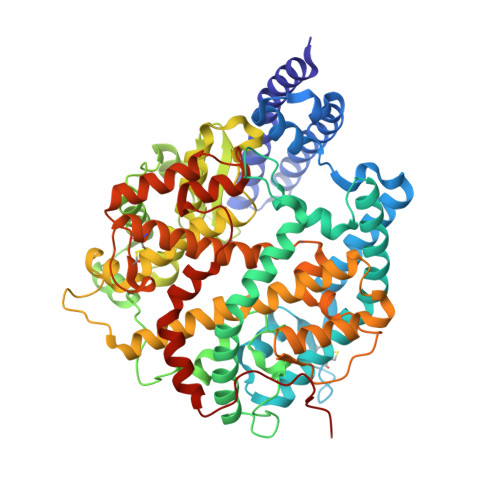 | |
UniProt | |||||
Find proteins for F6V9L3 (Equus caballus) Explore F6V9L3 Go to UniProtKB: F6V9L3 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | F6V9L3 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Spike protein S1 | 229 | Severe acute respiratory syndrome coronavirus 2 | Mutation(s): 0 Gene Names: S, 2 | 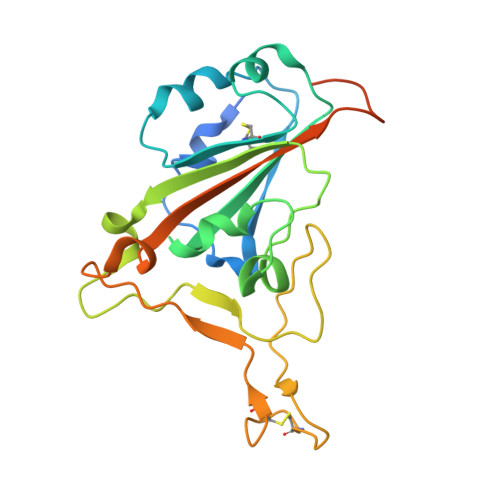 | |
UniProt | |||||
Find proteins for P0DTC2 (Severe acute respiratory syndrome coronavirus 2) Explore P0DTC2 Go to UniProtKB: P0DTC2 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0DTC2 | ||||
Glycosylation | |||||
| Glycosylation Sites: 1 | Go to GlyGen: P0DTC2-1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| NAG (Subject of Investigation/LOI) Query on NAG | D [auth B] | 2-acetamido-2-deoxy-beta-D-glucopyranose C8 H15 N O6 OVRNDRQMDRJTHS-FMDGEEDCSA-N |  | ||
| ZN (Subject of Investigation/LOI) Query on ZN | C [auth A] | ZINC ION Zn PTFCDOFLOPIGGS-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 195.57 | α = 90 |
| b = 195.57 | β = 90 |
| c = 149.77 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| xia2 | data reduction |
| xia2 | data scaling |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Natural Science Foundation of China (NSFC) | China | 32100752 |