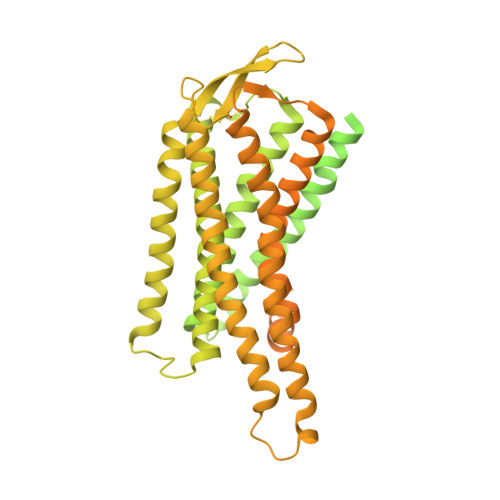
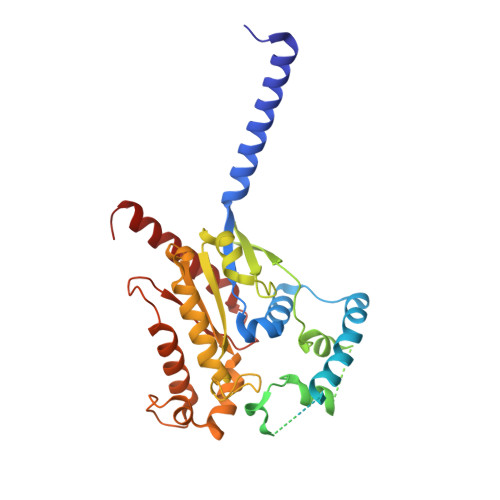
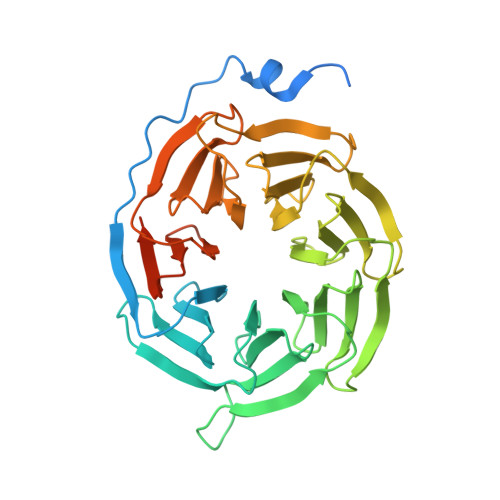
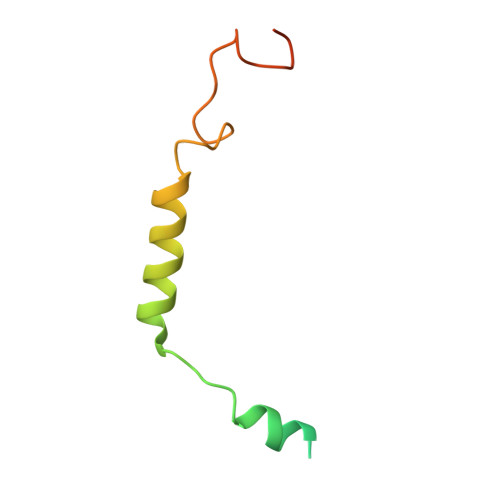
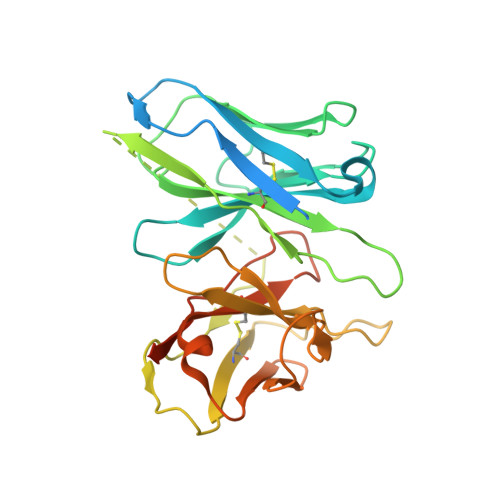
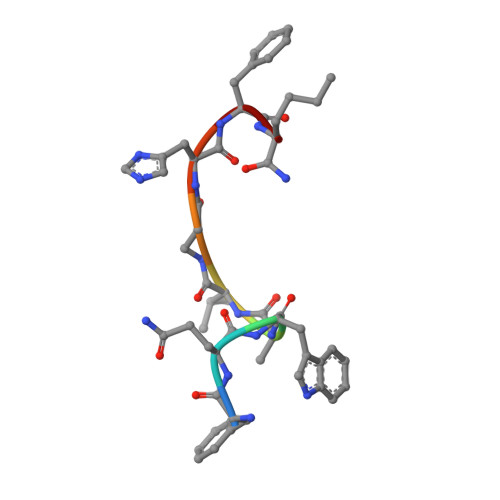
Structures of human gastrin-releasing peptide receptors bound to antagonist and agonist for cancer and itch therapy.
Peng, S., Zhan, Y., Zhang, D., Ren, L., Chen, A., Chen, Z.F., Zhang, H.(2023) Proc Natl Acad Sci U S A 120: e2216230120-e2216230120
- PubMed: 36724251
- DOI: https://doi.org/10.1073/pnas.2216230120
- Primary Citation of Related Structures:
7W3Z, 7W40, 7W41 - PubMed Abstract:
Gastrin releasing peptide receptor (GRPR), a member of the bombesin (BBN) G protein-coupled receptors, is aberrantly overexpressed in several malignant tumors, including those of the breast, prostate, pancreas, lung, and central nervous system. Additionally, it also mediates non-histaminergic itch and pathological itch conditions in mice. Thus, GRPR could be an attractive target for cancer and itch therapy. Here, we report the inactive state crystal structure of human GRPR in complex with the non-peptide antagonist PD176252, as well as two active state cryo-electron microscopy (cryo-EM) structures of GRPR bound to the endogenous peptide agonist gastrin-releasing peptide and the synthetic BBN analog [D-Phe 6 , β-Ala 11 , Phe 13 , Nle 14 ] Bn (6-14), in complex with G q heterotrimers. These structures revealed the molecular mechanisms for the ligand binding, receptor activation, and G q proteins signaling of GRPR, which are expected to accelerate the structure-based design of GRPR antagonists and agonists for the treatments of cancer and pruritus.
- Hangzhou Institute of Innovative Medicine, Institute of Pharmacology and Toxicology, Zhejiang Province Key Laboratory of Anti-Cancer Drug Research, College of Pharmaceutical Sciences, Zhejiang University, Hangzhou, Zhejiang 310058, China.
Organizational Affiliation: