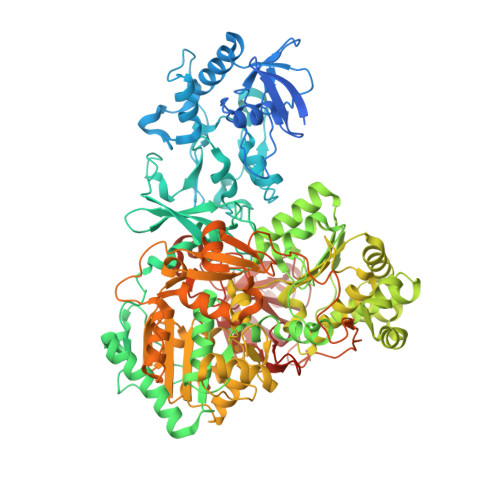
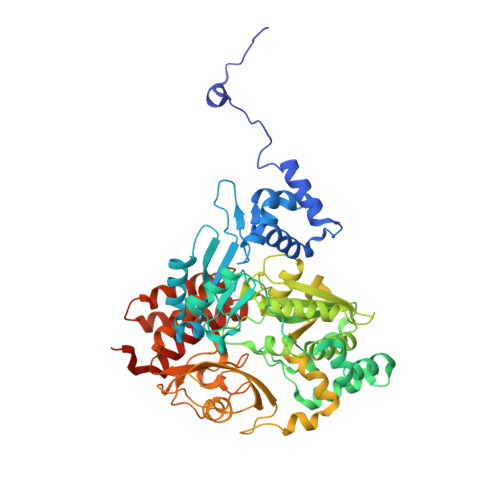
Multiple electron transfer pathways of tungsten-containing formate dehydrogenase in direct electron transfer-type bioelectrocatalysis.
Yoshikawa, T., Makino, F., Miyata, T., Suzuki, Y., Tanaka, H., Namba, K., Kano, K., Sowa, K., Kitazumi, Y., Shirai, O.(2022) Chem Commun (Camb) 58: 6478-6481
- PubMed: 35535582
- DOI: https://doi.org/10.1039/d2cc01541b
- Primary Citation of Related Structures:
7VW6 - PubMed Abstract:
Tungsten-containing formate dehydrogenase from Methylorubrum extroquens AM1 (FoDH1)-a promising biocatalyst for the interconversion of carbon dioxide/formate and nicotine adenine dinucleotide (NAD + )/NADH redox couples-was investigated using structural biology and bioelectrochemistry. FoDH1 is reported to be an enzyme that can realize "direct electron transfer (DET)-type bioelectrocatalysis." However, its 3-D structure, electrode-active sites, and electron transfer (ET) pathways remain unclear. The ET pathways were investigated using structural information, electrostatic interactions between the electrode and the enzyme, and the differences in the substrates. Two electrode-active sites and multiple ET pathways in FoDH1 were discovered.
- Division of Applied Life Sciences, Graduate School of Agriculture, Kyoto University, Sakyo Kyoto, 606-8502, Japan. sowa.keisei.2u@kyoto-u.ac.jp.
Organizational Affiliation: