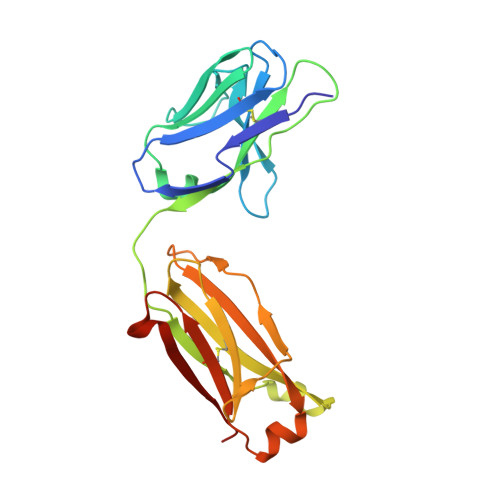
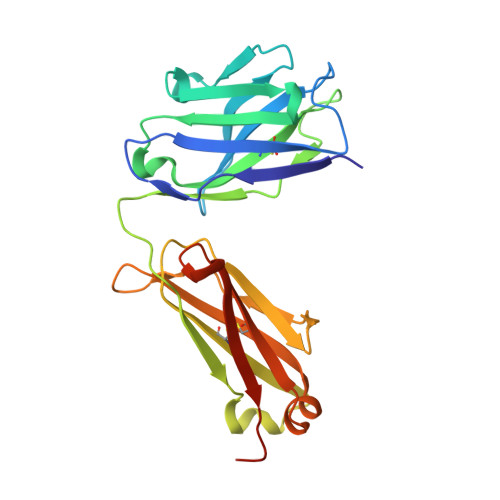
Design of a Novel Fab-Like Antibody Fragment with Enhanced Stability and Affinity for Clinical use.
Wang, C., Hong, J., Yang, Z., Zhou, X., Yang, Y., Kong, Y., Chen, B., Wu, H., Qian, B.Z., Dimitrov, D.S., Zhou, X., Wu, Y., Ying, T.(2022) Small Methods 6: e2100966-e2100966
- PubMed: 35174992
- DOI: https://doi.org/10.1002/smtd.202100966
- Primary Citation of Related Structures:
7VSU, 7VSW - PubMed Abstract:
With increasing interest in applying recombinant monoclonal antibodies (mAbs) in human medicine, engineered mAb fragments with reduced size and improved stability are in demand to overcome current limitations in clinical use. Herein, a novel Fab-like antibody fragment generated via an in silico-based engineering approach where the CH1 and CL domains of Fab are replaced by the IgG1 CH3 domains is described. This construct, designated as FabCH3, maintains the natural N-terminus and C-terminus of IgG antibody, can be expressed at a high level in bacterial cells and, importantly, exhibits much higher stability and affinity than the parental Fab when tested in a mesothelin-specific Fab m912, as well as a vascular endothelial growth factor A (VEGFA)-specific Fab Ranibizumab (in vivo). The high-resolution crystal structures of m912 FabCH3 and m912 Fab are determined, and the comparative analysis reveals more rigid structures in both constant domains and complementarity-determining regions of FabCH3, explaining its enhanced stability and affinity. Overall, the stabilized FabCH3 described in this report provides a versatile platform for engineering Fab-like antibody fragments with higher stability and antigen-binding affinity that can be used as a distinct class of antibody therapeutics.
- MOE/NHC Key Laboratory of Medical Molecular Virology, Shanghai Institute of Infectious Disease and Biosecurity, School of Basic Medical Sciences, Shanghai Medical College, Fudan University, Shanghai, 200032, China.
Organizational Affiliation: