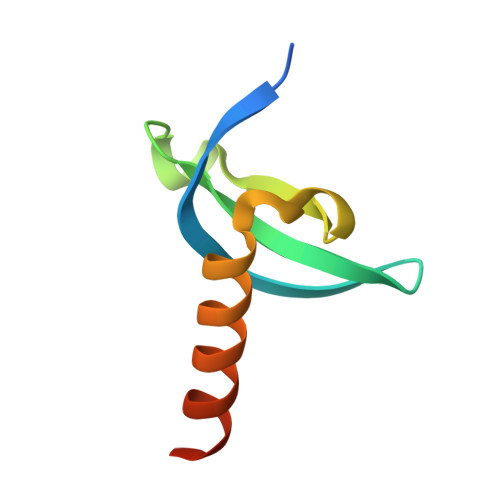
Structural insights into the chromodomain of Oxpecker in complex with histone H3 lysine 9 trimethylation reveal a transposon silencing mechanism by heterodimerization.
Jin, Z., Yu, B., Huang, Y.(2023) Biochem Biophys Res Commun 652: 95-102
- PubMed: 36841100
- DOI: https://doi.org/10.1016/j.bbrc.2023.02.045
- Primary Citation of Related Structures:
7VRF - PubMed Abstract:
Oxpecker, the homolog of Rhino/HP1D, exclusively expressed in Drosophila ovaries, belongs to the Heterochromatin Protein 1 family, as does Rhino. Rhi recognizes piRNA clusters enriched with the heterochromatin marker H3K9me3 via its N-terminal chromodomain and recruits Deadlock via its C-terminal chromoshadow domain, further recruits Moonshiner, a paralog of the TATA box-binding protein-related factor 2 large subunits, to promote transcription of piRNA precursors, thereby protecting the genome. Despite Oxp possessing only the chromodomain, its loss leads to the upregulation of transposons in the female germline. In this study, we solved the crystal structure of the Oxp chromodomain in complex with the histone H3K9me3 peptide. As the Oxp chromodomain dimerizes, two H3K9me3 peptides bind to the Oxp chromodomain in an antiparallel manner. ITC experiments and site-directed mutagenesis experiments showed that E44 determines Oxp's five-fold stronger binding ability to H3K9me3 than that of Rhi. In addition, we found that Oxp and Rhi can form a heterodimer, which may shed light on the molecular mechanism by which Oxp regulates transposon silencing in the absence of CSD.
- Department of General Surgery, Shanghai Key Laboratory of Biliary Tract Disease Research, State Key Laboratory of Oncogenes and Related Genes, Xinhua Hospital, Shanghai Jiao Tong University, Shanghai, 200092, China.
Organizational Affiliation: