Characterization of Multifunctional and Non-stereoselective Oxidoreductase RubE7/IstO, Expanding the Functional Diversity of the Flavoenzyme Superfamily.
Yan, Y., Yu, Z., Zhong, W., Hou, X., Tao, Q., Cao, M., Wang, L., Cai, X., Rao, Y., Huang, S.X.(2022) Angew Chem Int Ed Engl 61: e202200189-e202200189
- PubMed: 35191152
- DOI: https://doi.org/10.1002/anie.202200189
- Primary Citation of Related Structures:
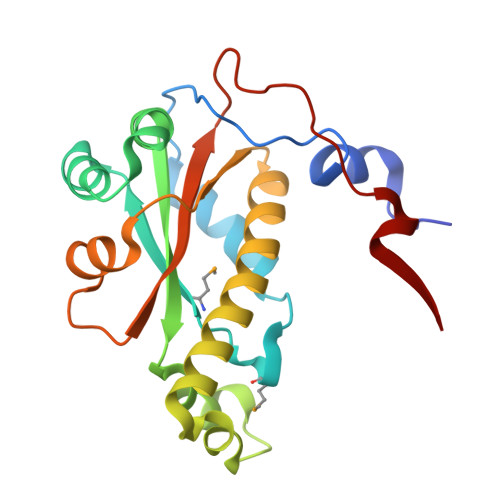
7VQK - PubMed Abstract:
Flavin-dependent enzymes enable a broad range of redox transformations and generally act as monofunctional and stereoselective catalysts. Herein, we report the investigation of a multifunctional and non-stereoselective FMN-dependent oxidoreductase RubE7 from the rubrolone biosynthetic pathway. Our study outlines a single RubE7-catalysed sequential reduction of three spatially distinct bonds in a tropolone ring and a reversible double-bond reduction and dehydrogenation. The crystal structure of IstO (a RubE7 homologue) with 2.0 Å resolution reveals the location of the active site at the interface of two monomers, and the size of active site is large enough to permit both flipping and free rotation of the substrate, resulting in multiple nonselective reduction reactions. Molecular docking and site mutation studies demonstrate that His106 is oriented towards the substrate and is important for the reverse dehydrogenation reaction.
- State Key Laboratory of Phytochemistry and Plant Resources in West China, CAS Center for Excellence in Molecular Plant Sciences, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming, 650201, China.
Organizational Affiliation: