Tumor suppressor BAP1 nuclear import is governed by transportin-1.
Yang, T.J., Li, T.N., Huang, R.S., Pan, M.Y., Lin, S.Y., Lin, S., Wu, K.P., Wang, L.H., Hsu, S.D.(2022) J Cell Biol 221
- PubMed: 35446349
- DOI: https://doi.org/10.1083/jcb.202201094
- Primary Citation of Related Structures:
7VPW - PubMed Abstract:
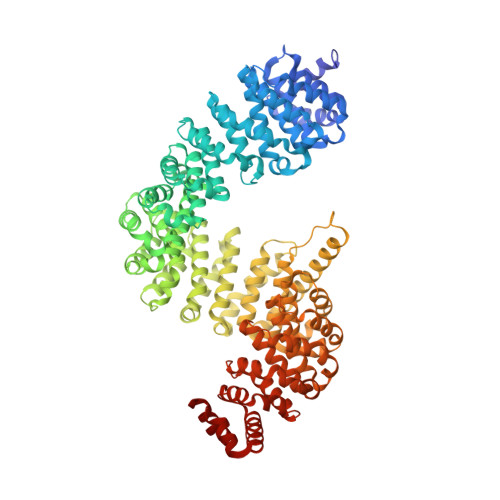
Subcellular localization of the deubiquitinating enzyme BAP1 is deterministic for its tumor suppressor activity. While the monoubiquitination of BAP1 by an atypical E2/E3-conjugated enzyme UBE2O and BAP1 auto-deubiquitination are known to regulate its nuclear localization, the molecular mechanism by which BAP1 is imported into the nucleus has remained elusive. Here, we demonstrated that transportin-1 (TNPO1, also known as Karyopherin β2 or Kapβ2) targets an atypical C-terminal proline-tyrosine nuclear localization signal (PY-NLS) motif of BAP1 and serves as the primary nuclear transporter of BAP1 to achieve its nuclear import. TNPO1 binding dissociates dimeric BAP1 and sequesters the monoubiquitination sites flanking the PY-NLS of BAP1 to counteract the function of UBE2O that retains BAP1 in the cytosol. Our findings shed light on how TNPO1 regulates the nuclear import, self-association, and monoubiquitination of BAP1 pertinent to oncogenesis.
- Institute of Biological Chemistry, Academia Sinica, Taipei, Taiwan.
Organizational Affiliation: