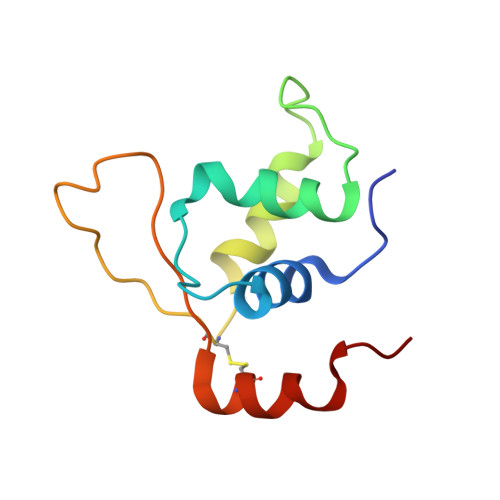
FOXL2 and FOXA1 cooperatively assemble on the TP53 promoter in alternative dimer configurations.
Choi, Y., Luo, Y., Lee, S., Jin, H., Yoon, H.J., Hahn, Y., Bae, J., Lee, H.H.(2022) Nucleic Acids Res 50: 8929-8946
- PubMed: 35920317
- DOI: https://doi.org/10.1093/nar/gkac673
- Primary Citation of Related Structures:
7VOU, 7VOV, 7VOX - PubMed Abstract:
Although both the p53 and forkhead box (FOX) family proteins are key transcription factors associated with cancer progression, their direct relationship is unknown. Here, we found that FOX family proteins bind to the non-canonical homotypic cluster of the p53 promoter region (TP53). Analysis of crystal structures of FOX proteins (FOXL2 and FOXA1) bound to the p53 homotypic cluster indicated that they interact with a 2:1 stoichiometry accommodated by FOX-induced DNA allostery. In particular, FOX proteins exhibited distinct dimerization patterns in recognition of the same p53-DNA; dimer formation of FOXA1 involved protein-protein interaction, but FOXL2 did not. Biochemical and biological functional analyses confirmed the cooperative binding of FOX proteins to the TP53 promoter for the transcriptional activation of TP53. In addition, up-regulation of TP53 was necessary for FOX proteins to exhibit anti-proliferative activity in cancer cells. These analyses reveal the presence of a discrete characteristic within FOX family proteins in which FOX proteins regulate the transcription activity of the p53 tumor suppressor via cooperative binding to the TP53 promoter in alternative dimer configurations.
- Department of Chemistry, College of Natural Sciences, Seoul National University, Seoul 08826, Korea.
Organizational Affiliation: