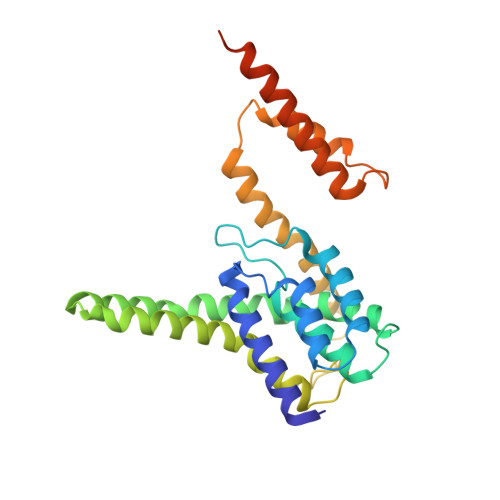
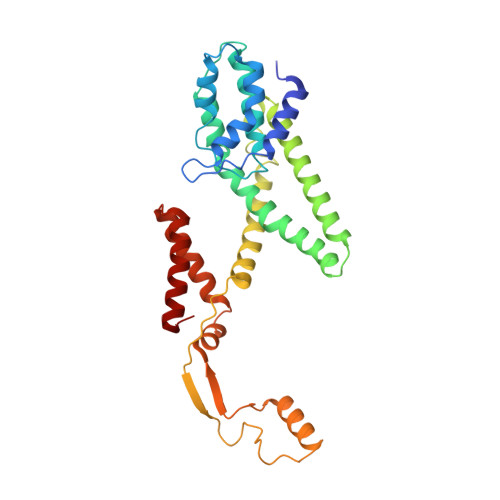
Structural Basis of Pore Formation in the Mannose Phosphotransferase System by Pediocin PA-1.
Zhu, L., Zeng, J., Wang, C., Wang, J.(2022) Appl Environ Microbiol 88: e0199221-e0199221
- PubMed: 34851716
- DOI: https://doi.org/10.1128/AEM.01992-21
- Primary Citation of Related Structures:
7VLX, 7VLY - PubMed Abstract:
Bacteriocins are ribosomally synthesized bacterial antimicrobial peptides that have a narrow spectrum of antibacterial activity against species closely related to the producers. Pediocin-like (or class IIa) bacteriocins (PLBs) exhibit antibacterial activity against several Gram-positive bacterial strains by forming pores in the cytoplasmic membrane of target cells with a specific receptor, the mannose phosphotransferase system (man-PTS). In this study, we report the cryo-electron microscopy structures of man-PTS from Listeria monocytogenes alone and its complex with pediocin PA-1, the first and most extensively studied representative PLB, at resolutions of 3.12 and 2.45 Å, respectively. The structures revealed that the binding of pediocin PA-1 opens the Core domain of man-PTS away from its Vmotif domain, creating a pore through the cytoplasmic membranes of target cells. During this process, the N-terminal β-sheet region of pediocin PA-1 can specifically attach to the extracellular surface of the man-PTS Core domain, whereas the C-terminal half penetrates the membrane and cracks the man-PTS like a wedge. Thus, our findings shed light on a design of novel PLBs that can kill the target pathogenic bacteria. IMPORTANCE Listeria monocytogenes is a ubiquitous microorganism responsible for listeriosis, a rare but severe disease in humans, who become infected by ingesting contaminated food products (i.e., dairy, meat, fish, and vegetables): the disease has a fatality rate of 33%. Pediocin PA-1 is an important commercial additive used in food production to inhibit Listeria species. The mannose phosphotransferase system (man-PTS) is responsible for the sensitivity of Listeria monocytogenes to pediocin PA-1. In this study, we report the cryo-EM structures of man-PTS from Listeria monocytogenes alone and its complex with pediocin PA-1 at resolutions of 3.12 and 2.45 Å, respectively. Our results facilitate the understanding of the mode of action of class IIa bacteriocins as an alternative to antibiotics.
- State Key Laboratory of Membrane Biology, Beijing Advanced Innovation Center for Structural Biology, School of Life Sciences, Tsinghua University, Beijing, People's Republic of China.
Organizational Affiliation: