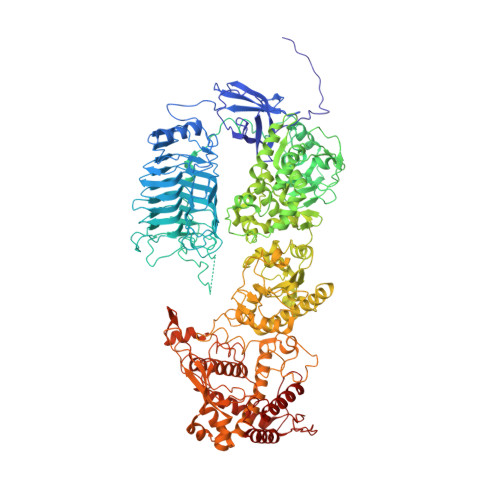
A substrate-induced gating mechanism is conserved among Gram-positive IgA1 metalloproteases.
Redzic, J.S., Rahkola, J., Tran, N., Holyoak, T., Lee, E., Martin-Galiano, A.J., Meyer, N., Zheng, H., Eisenmesser, E.(2022) Commun Biol 5: 1190-1190
- PubMed: 36336763
- DOI: https://doi.org/10.1038/s42003-022-04173-3
- Primary Citation of Related Structures:
7UVK, 7UVL - PubMed Abstract:
The mucosal adaptive immune response is dependent on the production of IgA antibodies and particularly IgA1, yet opportunistic bacteria have evolved mechanisms to specifically block this response by producing IgA1 proteases (IgA1Ps). Our lab was the first to describe the structures of a metal-dependent IgA1P (metallo-IgA1P) produced from Gram-positive Streptococcus pneumoniae both in the absence and presence of its IgA1 substrate through cryo-EM single particle reconstructions. This prior study revealed an active-site gating mechanism reliant on substrate-induced conformational changes to the enzyme that begged the question of whether such a mechanism is conserved among the wider Gram-positive metallo-IgA1P subfamily of virulence factors. Here, we used cryo-EM to characterize the metallo-IgA1P of a more distantly related family member from Gemella haemolysans, an emerging opportunistic pathogen implicated in meningitis, endocarditis, and more recently bacteremia in the elderly. While the substrate-free structures of these two metallo-IgA1Ps exhibit differences in the relative starting positions of the domain responsible for gating substrate, the enzymes have similar domain orientations when bound to IgA1. Together with biochemical studies that indicate these metallo-IgA1Ps have similar binding affinities and activities, these data indicate that metallo-IgA1P binding requires the specific IgA1 substrate to open the enzymes for access to their active site and thus, largely conform to an "induced fit" model.
- Department of Biochemistry and Molecular Genetics, School of Medicine, University of Colorado Denver, School of Medicine, Aurora, CO, 80045, USA.
Organizational Affiliation: