Structures of the TMC-1 complex illuminate mechanosensory transduction.
Jeong, H., Clark, S., Goehring, A., Dehghani-Ghahnaviyeh, S., Rasouli, A., Tajkhorshid, E., Gouaux, E.(2022) Nature 610: 796-803
- PubMed: 36224384
- DOI: https://doi.org/10.1038/s41586-022-05314-8
- Primary Citation of Related Structures:
7USW, 7USX, 7USY - PubMed Abstract:
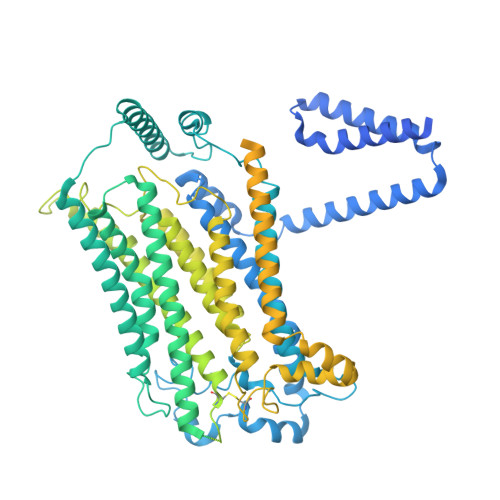
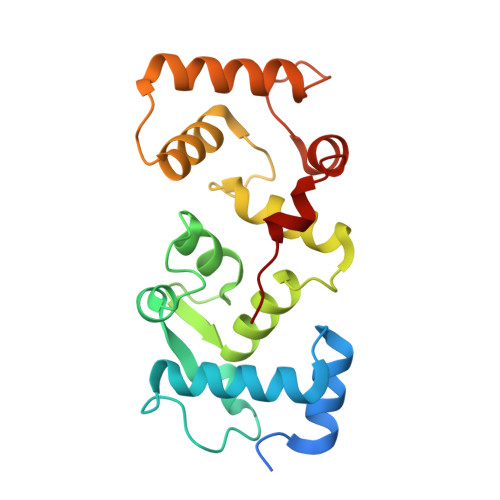
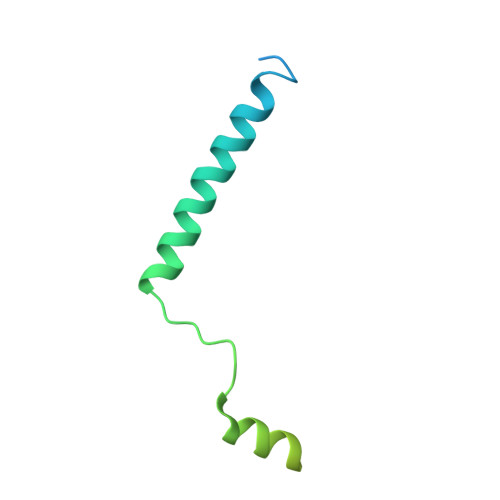
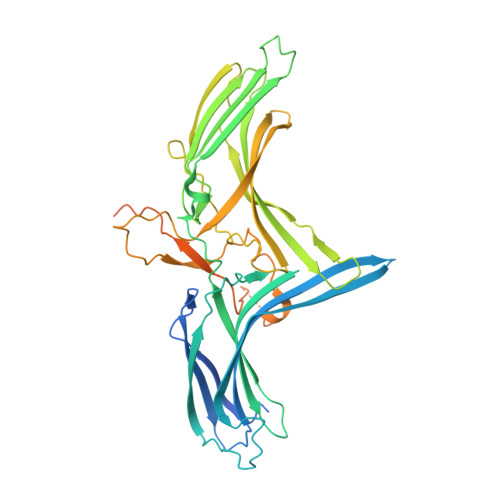
The initial step in the sensory transduction pathway underpinning hearing and balance in mammals involves the conversion of force into the gating of a mechanosensory transduction channel 1 . Despite the profound socioeconomic impacts of hearing disorders and the fundamental biological significance of understanding mechanosensory transduction, the composition, structure and mechanism of the mechanosensory transduction complex have remained poorly characterized. Here we report the single-particle cryo-electron microscopy structure of the native transmembrane channel-like protein 1 (TMC-1) mechanosensory transduction complex isolated from Caenorhabditis elegans. The two-fold symmetric complex is composed of two copies each of the pore-forming TMC-1 subunit, the calcium-binding protein CALM-1 and the transmembrane inner ear protein TMIE. CALM-1 makes extensive contacts with the cytoplasmic face of the TMC-1 subunits, whereas the single-pass TMIE subunits reside on the periphery of the complex, poised like the handles of an accordion. A subset of complexes additionally includes a single arrestin-like protein, arrestin domain protein (ARRD-6), bound to a CALM-1 subunit. Single-particle reconstructions and molecular dynamics simulations show how the mechanosensory transduction complex deforms the membrane bilayer and suggest crucial roles for lipid-protein interactions in the mechanism by which mechanical force is transduced to ion channel gating.
- Vollum Institute, Oregon Health and Science University, Portland, OR, USA.
Organizational Affiliation: