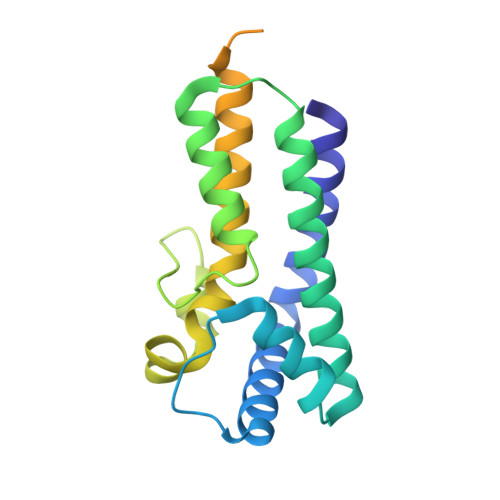
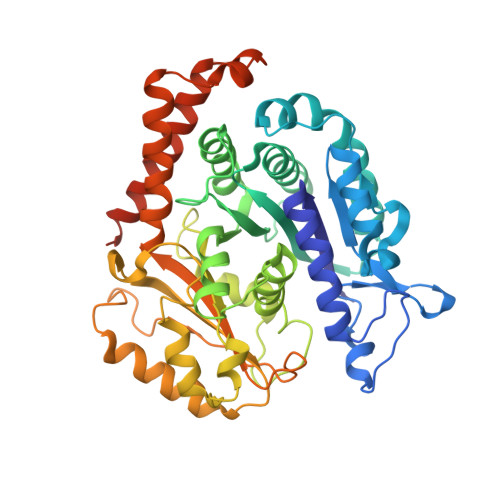
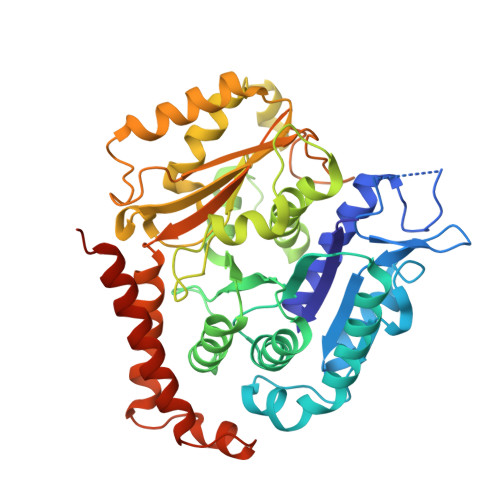
SPACA9 is a lumenal protein of human ciliary singlet and doublet microtubules.
Gui, M., Croft, J.T., Zabeo, D., Acharya, V., Kollman, J.M., Burgoyne, T., Hoog, J.L., Brown, A.(2022) Proc Natl Acad Sci U S A 119: e2207605119-e2207605119
- PubMed: 36191189
- DOI: https://doi.org/10.1073/pnas.2207605119
- Primary Citation of Related Structures:
7UN1, 7UNG - PubMed Abstract:
The cilium-centrosome complex contains triplet, doublet, and singlet microtubules. The lumenal surfaces of each microtubule within this diverse array are decorated by microtubule inner proteins (MIPs). Here, we used single-particle cryo-electron microscopy methods to build atomic models of two types of human ciliary microtubule: the doublet microtubules of multiciliated respiratory cells and the distal singlet microtubules of monoflagellated human spermatozoa. We discover that SPACA9 is a polyspecific MIP capable of binding both microtubule types. SPACA9 forms intralumenal striations in the B tubule of respiratory doublet microtubules and noncontinuous spirals in sperm singlet microtubules. By acquiring new and reanalyzing previous cryo-electron tomography data, we show that SPACA9-like intralumenal striations are common features of different microtubule types in animal cilia. Our structures provide detailed references to help rationalize ciliopathy-causing mutations and position cryo-EM as a tool for the analysis of samples obtained directly from ciliopathy patients.
- Department of Biological Chemistry and Molecular Pharmacology, Harvard Medical School, Boston, MA 02115.
Organizational Affiliation: