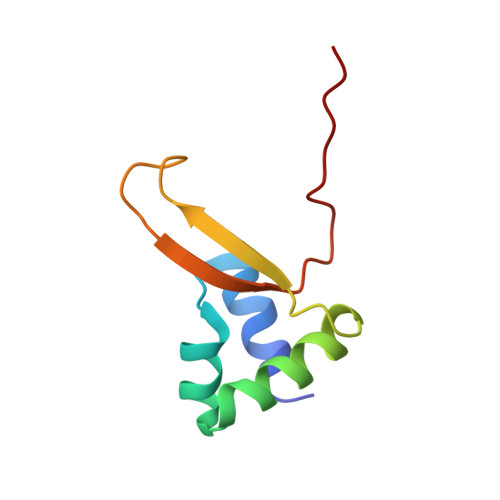
The structure of Vibrio cholerae FeoC reveals conservation of the helix-turn-helix motif but not the cluster-binding domain.
Brown, J.B., Lee, M.A., Smith, A.T.(2022) J Biol Inorg Chem 27: 485-495
- PubMed: 35796835
- DOI: https://doi.org/10.1007/s00775-022-01945-4
- Primary Citation of Related Structures:
7U37 - PubMed Abstract:
Most pathogenic bacteria require ferrous iron (Fe 2+ ) in order to sustain infection within hosts. The ferrous iron transport (Feo) system is the most highly conserved prokaryotic transporter of Fe 2+ , but its mechanism remains to be fully characterized. Most Feo systems are composed of two proteins: FeoA, a soluble SH3-like accessory protein, and FeoB, a membrane protein that translocates Fe 2+ across a lipid bilayer. Some bacterial feo operons encode FeoC, a third soluble, winged-helix protein that remains enigmatic in function. We previously demonstrated that selected FeoC proteins bind O 2 -sensitive [4Fe-4S] clusters via Cys residues, leading to the proposal that some FeoCs could sense O 2 to regulate Fe 2+ transport. However, not all FeoCs conserve these Cys residues, and FeoC from the causative agent of cholera (Vibrio cholerae) notably lacks any Cys residues, precluding cluster binding. In this work, we determined the NMR structure of VcFeoC, which is monomeric and conserves the helix-turn-helix domain seen in other FeoCs. In contrast, however, the structure of VcFeoC reveals a truncated winged β-sheet in which the cluster-binding domain is notably absent. Using homology modeling, we predicted the structure of VcNFeoB and used docking to identify an interaction site with VcFeoC, which is confirmed by NMR spectroscopy. These findings provide the first atomic-level structure of VcFeoC and contribute to a better understanding of its role vis-à-vis FeoB.
- Department of Chemistry and Biochemistry, University of Maryland, Baltimore County, Baltimore, MD, 21250, USA.
Organizational Affiliation: