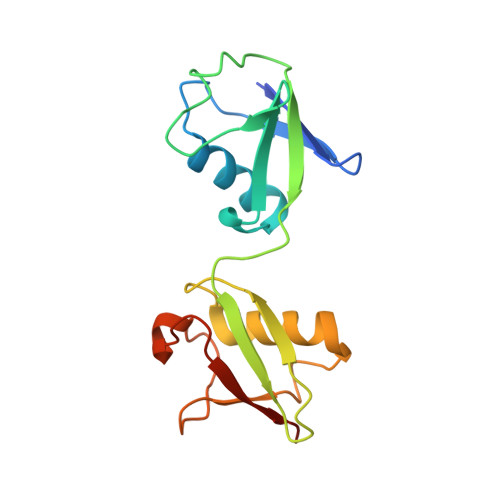
Structural basis for the simultaneous recognition of NEMO and acceptor ubiquitin by the HOIP NZF1 domain.
Rahighi, S., Iyer, M., Oveisi, H., Nasser, S., Duong, V.(2022) Sci Rep 12: 12241-12241
- PubMed: 35851409
- DOI: https://doi.org/10.1038/s41598-022-16193-4
- Primary Citation of Related Structures:
7TV4 - PubMed Abstract:
Ubiquitination of NEMO by the linear ubiquitin chain assembly complex (LUBAC) is essential for activating the canonical NF-κB signaling pathway. While the NZF1 domain of the HOIP subunit of LUBAC recognizes the NEMO substrate, it is unclear how it cooperates with the catalytic domains in the ubiquitination process. Here, we report a crystal structure of NEMO in complex with HOIP NZF1 and linear diubiquitin chains, in which the two proteins bind to distinct sites on NEMO. Moreover, the NZF1 domain simultaneously interacts with NEMO and Ile44 surface of a proximal ubiquitin from a linear diubiquitin chain, where the C-term tail of the ubiquitin is in the proximity of the NEMO ubiquitination site (Lys285). We further propose a model for the linear ubiquitination of NEMO by HOIP. In the model, NZF1 binds the monoubiquitinated NEMO and recruits the catalytic domains to the ubiquitination site, thereby ensuring site-specific ubiquitination of NEMO.
- Chapman University School of Pharmacy (CUSP), Harry and Diane Rinker Health Science Campus, Chapman University, Irvine, CA, 92618, USA. rahighi@chapman.edu.
Organizational Affiliation: