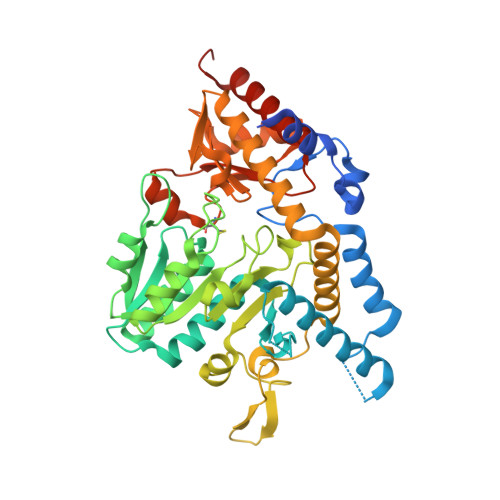
Structural analysis of Atopobium parvulum SufS cysteine desulfurase linked to Crohn's disease.
Karunakaran, G., Yang, Y., Tremblay, V., Ning, Z., Martin, J., Belaouad, A., Figeys, D., Brunzelle, J.S., Giguere, P.M., Stintzi, A., Couture, J.F.(2022) FEBS Lett 596: 898-909
- PubMed: 35122247
- DOI: https://doi.org/10.1002/1873-3468.14295
- Primary Citation of Related Structures:
7TLM, 7TLP, 7TLQ, 7TLR - PubMed Abstract:
Crohn's disease (CD) is characterized by the chronic inflammation of the gastrointestinal tract. A dysbiotic microbiome and a defective immune system are linked to CD, where hydrogen sulfide (H 2 S) microbial producers positively correlate with the severity of the disease. Atopobium parvulum is a key H 2 S producer from the microbiome of CD patients. In this study, the biochemical characterization of two Atopobium parvulum cysteine desulfurases, ApSufS and ApCsdB, shows that the enzymes are allosterically regulated. Structural analyses reveal that ApSufS forms a dimer with conserved characteristics observed in type II cysteine desulfurases. Four residues surrounding the active site are essential to catalyse cysteine desulfurylation, and a segment of short-chain residues grant access for substrate binding. A better understanding of ApSufS will help future avenues for CD treatment.
- Department of Biochemistry, Microbiology and Immunology, Faculty of Medicine, University of Ottawa, Canada.
Organizational Affiliation: