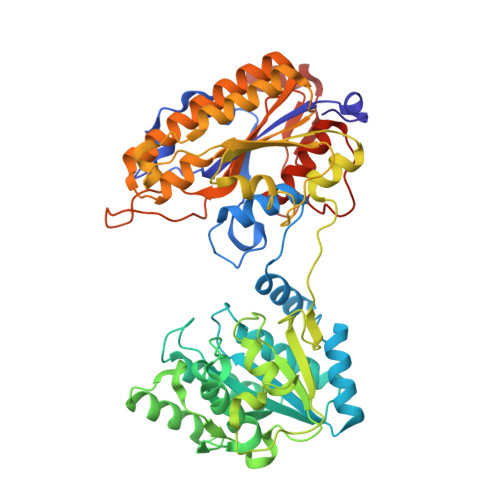
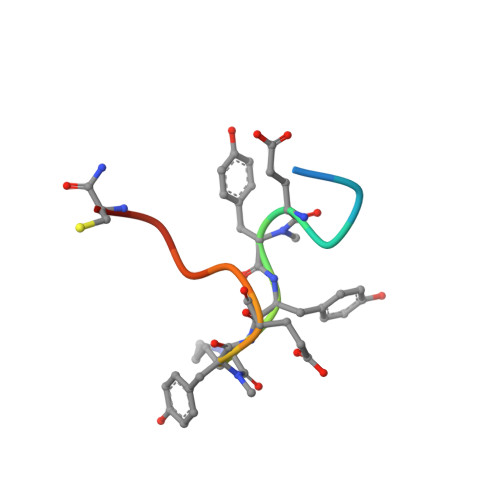
Serum-Stable and Selective Backbone-N-Methylated Cyclic Peptides That Inhibit Prokaryotic Glycolytic Mutases.
van Neer, R.H.P., Dranchak, P.K., Liu, L., Aitha, M., Queme, B., Kimura, H., Katoh, T., Battaile, K.P., Lovell, S., Inglese, J., Suga, H.(2022) ACS Chem Biol 17: 2284-2295
- PubMed: 35904259
- DOI: https://doi.org/10.1021/acschembio.2c00403
- Primary Citation of Related Structures:
7TL7, 7TL8 - PubMed Abstract:
N -Methylated amino acids ( N -MeAAs) are privileged residues of naturally occurring peptides critical to bioactivity. However, de novo discovery from ribosome display is limited by poor incorporation of N -methylated amino acids into the nascent peptide chain attributed to a poor EF-Tu affinity for the N -methyl-aminoacyl-tRNA. By reconfiguring the tRNA's T-stem region to compensate and tune the EF-Tu affinity, we conducted Random nonstandard Peptides Integrated Discovery (RaPID) display of a macrocyclic peptide (MCP) library containing six different N -MeAAs. We have here devised a "pool-and-split" enrichment strategy using the RaPID display and identified N -methylated MCPs against three species of prokaryotic metal-ion-dependent phosphoglycerate mutases. The enriched MCPs reached 57% N -methylation with up to three consecutively incorporated N -MeAAs, rivaling natural products. Potent nanomolar inhibitors ranging in ortholog selectivity, strongly mediated by N -methylation, were identified. Co-crystal structures reveal an architecturally related Ce-2 Ipglycermide active-site metal-ion-coordinating Cys lariat MCP, functionally dependent on two cis N -MeAAs with broadened iPGM species selectivity over the original nematode-selective MCPs. Furthermore, the isolation of a novel metal-ion-independent Staphylococcus aureus iPGM inhibitor utilizing a phosphoglycerate mimetic mechanism illustrates the diversity of possible chemotypes encoded by the N -MeAA MCP library.
- Department of Chemistry, Graduate School of Science, The University of Tokyo, Tokyo 113-0033, Japan.
Organizational Affiliation: