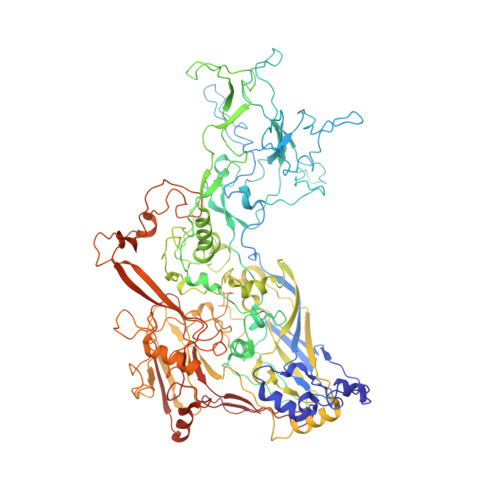
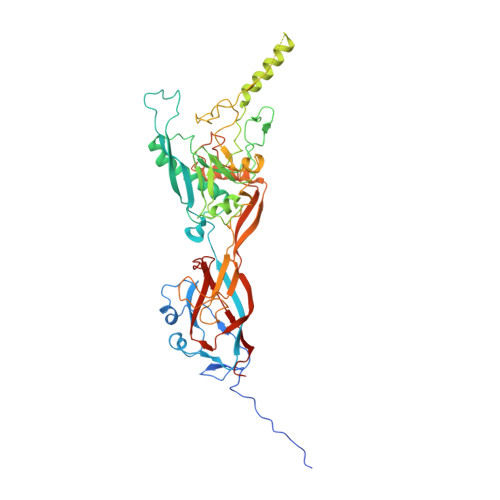
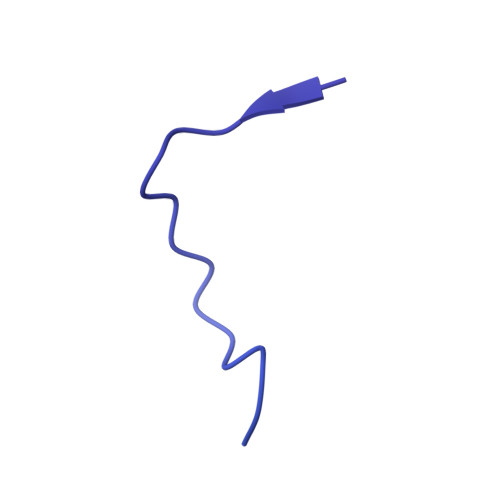
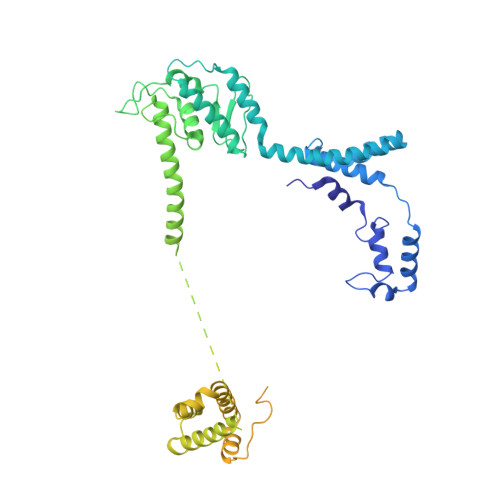
Refined Capsid Structure of Human Adenovirus D26 at 3.4 angstrom Resolution.
Reddy, V.S., Yu, X., Barry, M.A.(2022) Viruses 14
- PubMed: 35216007
- DOI: https://doi.org/10.3390/v14020414
- Primary Citation of Related Structures:
7TAU - PubMed Abstract:
Various adenoviruses are being used as viral vectors for the generation of vaccines against chronic and emerging diseases (e.g., AIDS, COVID-19). Here, we report the improved capsid structure for one of these vectors, human adenovirus D26 (HAdV-D26), at 3.4 Å resolution, by reprocessing the previous cryo-electron microscopy dataset and obtaining a refined model. In addition to overall improvements in the model, the highlights of the structure include (1) locating a segment of the processed peptide of VIII that was previously believed to be released from the mature virions, (2) reorientation of the helical appendage domain (APD) of IIIa situated underneath the vertex region relative to its counterpart observed in the cleavage defective ( ts1 ) mutant of HAdV-C5 that resulted in the loss of interactions between the APD and hexon bases, and (3) the revised conformation of the cleaved N-terminal segments of pre-protein VI (pVIn), located in the hexon cavities, is highly conserved, with notable stacking interactions between the conserved His13 and Phe18 residues. Taken together, the improved model of HAdV-D26 capsid provides a better understanding of protein-protein interactions in HAdV capsids and facilitates the efforts to modify and/or design adenoviral vectors with altered properties. Last but not least, we provide some insights into clotting factors (e.g., FX and PF4) binding to AdV vectors.
- Department of Integrative Structural and Computational Biology, The Scripps Research Institute, La Jolla, CA 92037, USA.
Organizational Affiliation: