Cryo-EM structure of CH235.12 in complex with HIV-1 Env trimer CH505TF.N279K.G458Y.SOSIP.664
Manne, K.To be published.
Experimental Data Snapshot
Starting Models: experimental
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Envelope glycoprotein gp160 | 461 | Human immunodeficiency virus 1 | Mutation(s): 0 Gene Names: env | 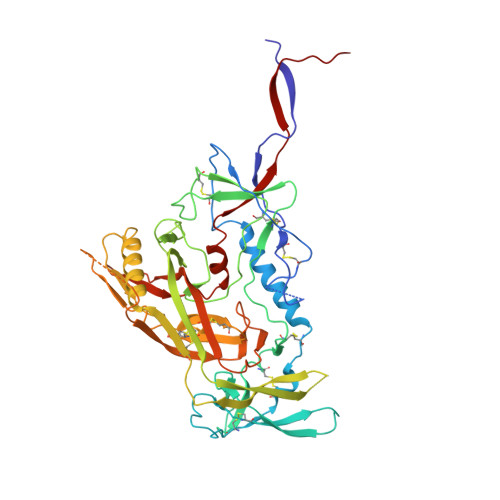 | |
UniProt | |||||
Find proteins for M4M0W3 (Human immunodeficiency virus type 1) Explore M4M0W3 Go to UniProtKB: M4M0W3 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | M4M0W3 | ||||
Glycosylation | |||||
| Glycosylation Sites: 8 | |||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Envelope glycoprotein gp41 | 160 | Human immunodeficiency virus 1 | Mutation(s): 0 Gene Names: env | 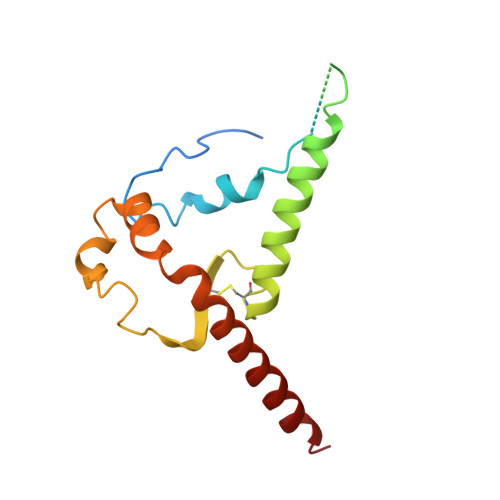 | |
UniProt | |||||
Find proteins for Q2N0S5 (Human immunodeficiency virus type 1) Explore Q2N0S5 Go to UniProtKB: Q2N0S5 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q2N0S5 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| CH235.12 Fab Heavy Chain | 225 | Homo sapiens | Mutation(s): 0 | 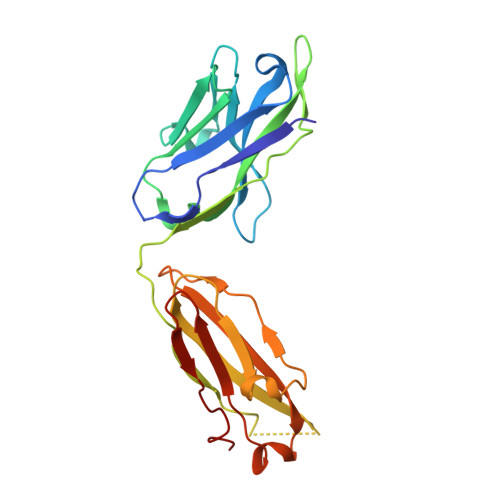 | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| CH235.12 Fab Light Chain | 213 | Homo sapiens | Mutation(s): 0 | 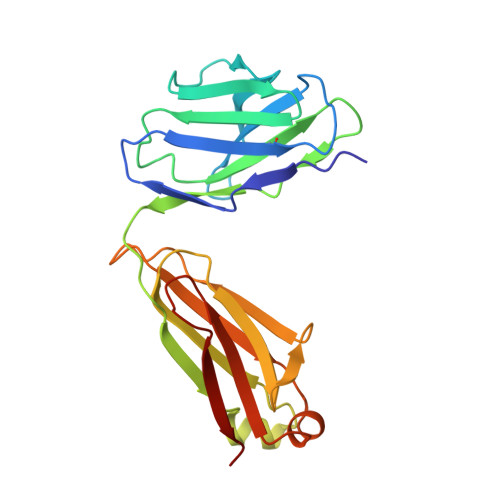 | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 5 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | 2D Diagram | Glycosylation | 3D Interactions |
| 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | M [auth a], S [auth O], Y [auth U] | 4 |  | N-Glycosylation | |
Glycosylation Resources | |||||
GlyTouCan: G95663PY GlyCosmos: G95663PY GlyGen: G95663PY | |||||
Entity ID: 6 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | 2D Diagram | Glycosylation | 3D Interactions |
| 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | DA [auth Z], N [auth b], R [auth N], T [auth P], X [auth T], DA [auth Z], N [auth b], R [auth N], T [auth P], X [auth T], Z [auth V] | 2 |  | N-Glycosylation | |
Glycosylation Resources | |||||
GlyTouCan: G42666HT GlyCosmos: G42666HT GlyGen: G42666HT | |||||
Entity ID: 7 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | 2D Diagram | Glycosylation | 3D Interactions |
| beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | AA [auth W], CA [auth Y], O [auth c], Q [auth M], U [auth Q], AA [auth W], CA [auth Y], O [auth c], Q [auth M], U [auth Q], W [auth S] | 3 |  | N-Glycosylation | |
Glycosylation Resources | |||||
GlyTouCan: G15407YE GlyCosmos: G15407YE GlyGen: G15407YE | |||||
Entity ID: 8 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | 2D Diagram | Glycosylation | 3D Interactions |
| alpha-D-mannopyranose-(1-6)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | BA [auth X], P [auth d], V [auth R] | 6 |  | N-Glycosylation | |
Glycosylation Resources | |||||
GlyTouCan: G34442SS GlyCosmos: G34442SS GlyGen: G34442SS | |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| NAG (Subject of Investigation/LOI) Query on NAG | EA [auth A] FA [auth A] GA [auth E] HA [auth E] IA [auth I] | 2-acetamido-2-deoxy-beta-D-glucopyranose C8 H15 N O6 OVRNDRQMDRJTHS-FMDGEEDCSA-N |  | ||
| Task | Software Package | Version |
|---|---|---|
| MODEL REFINEMENT | Coot | |
| MODEL REFINEMENT | PHENIX | |
| MODEL REFINEMENT | ISOLDE | |
| RECONSTRUCTION | cryoSPARC |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Institutes of Health/National Institute Of Allergy and Infectious Diseases (NIH/NIAID) | United States | AI45687 |