Two diphosphorylated degrons control c-Myc degradation by the Fbw7 tumor suppressor.
Welcker, M., Wang, B., Rusnac, D.V., Hussaini, Y., Swanger, J., Zheng, N., Clurman, B.E.(2022) Sci Adv 8: eabl7872-eabl7872
- PubMed: 35089787
- DOI: https://doi.org/10.1126/sciadv.abl7872
- Primary Citation of Related Structures:
7T1Y, 7T1Z - PubMed Abstract:
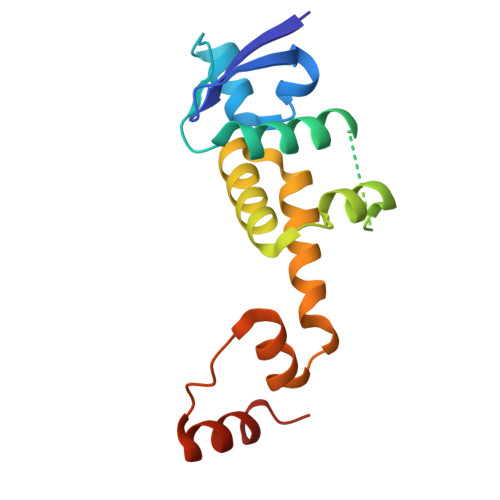
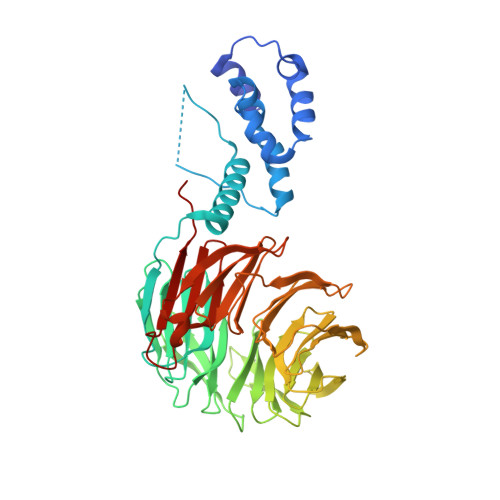
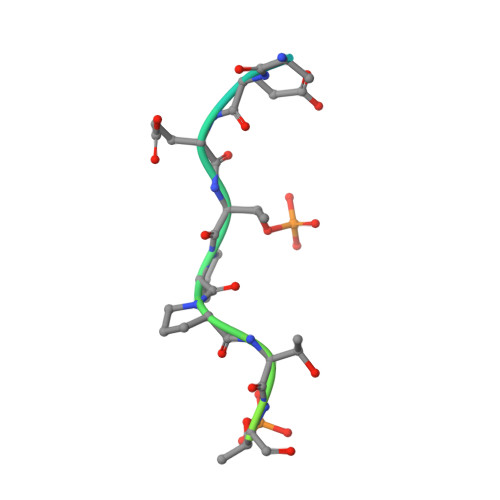
c-Myc (hereafter, Myc) is a cancer driver whose abundance is regulated by the SCF Fbw7 ubiquitin ligase and proteasomal degradation. Fbw7 binds to a phosphorylated Myc degron centered at threonine 58 (T58), and mutations of Fbw7 or T58 impair Myc degradation in cancers. Here, we identify a second Fbw7 phosphodegron at Myc T244 that is required for Myc ubiquitylation and acts in concert with T58 to engage Fbw7. While Ras-dependent Myc serine 62 phosphorylation (pS62) is thought to stabilize Myc by preventing Fbw7 binding, we find instead that pS62 greatly enhances Fbw7 binding and is an integral part of a high-affinity degron. Crystallographic studies revealed that both degrons bind Fbw7 in their diphosphorylated forms and that the T244 degron is recognized via a unique mode involving Fbw7 arginine 689 (R689), a mutational hotspot in cancers. These insights have important implications for Myc-associated tumorigenesis and therapeutic strategies targeting Myc stability.
- Clinical Research Division, Fred Hutchinson Cancer Research Center, Seattle, WA 98109, USA.
Organizational Affiliation: