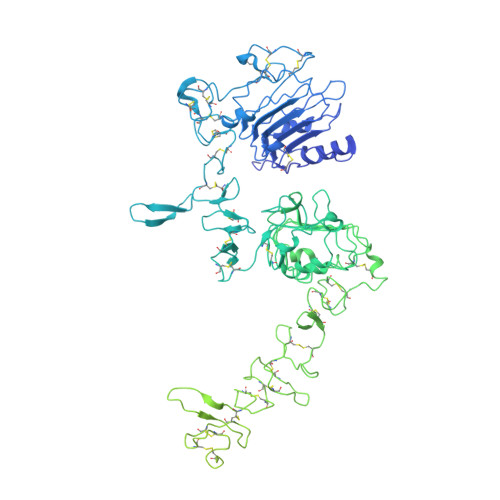
A molecular mechanism for the generation of ligand-dependent differential outputs by the epidermal growth factor receptor.
Huang, Y., Ognjenovic, J., Karandur, D., Miller, K., Merk, A., Subramaniam, S., Kuriyan, J.(2021) Elife 10
- PubMed: 34846302
- DOI: https://doi.org/10.7554/eLife.73218
- Primary Citation of Related Structures:
7SYD, 7SYE, 7SZ0, 7SZ1, 7SZ5, 7SZ7 - PubMed Abstract:
The epidermal growth factor receptor (EGFR) is a receptor tyrosine kinase that couples the binding of extracellular ligands, such as EGF and transforming growth factor-α (TGF-α), to the initiation of intracellular signaling pathways. EGFR binds to EGF and TGF-α with similar affinity, but generates different signals from these ligands. To address the mechanistic basis of this phenomenon, we have carried out cryo-EM analyses of human EGFR bound to EGF and TGF-α. We show that the extracellular module adopts an ensemble of dimeric conformations when bound to either EGF or TGF-α. The two extreme states of this ensemble represent distinct ligand-bound quaternary structures in which the membrane-proximal tips of the extracellular module are either juxtaposed or separated. EGF and TGF-α differ in their ability to maintain the conformation with the membrane-proximal tips of the extracellular module separated, and this conformation is stabilized preferentially by an oncogenic EGFR mutation. Close proximity of the transmembrane helices at the junction with the extracellular module has been associated previously with increased EGFR activity. Our results show how EGFR can couple the binding of different ligands to differential modulation of this proximity, thereby suggesting a molecular mechanism for the generation of ligand-sensitive differential outputs in this receptor family.
- Department of Molecular and Cell Biology, University of California, Berkeley, Berkeley, United States.
Organizational Affiliation: