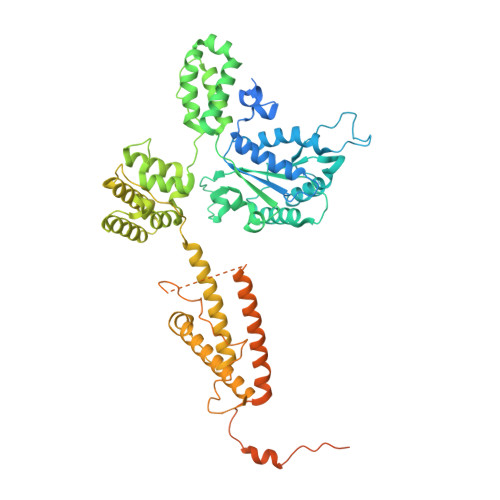
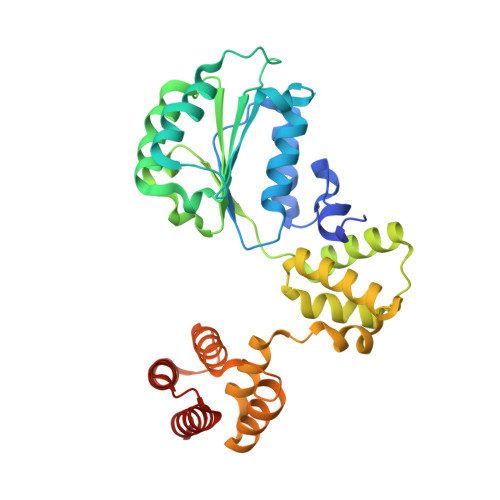
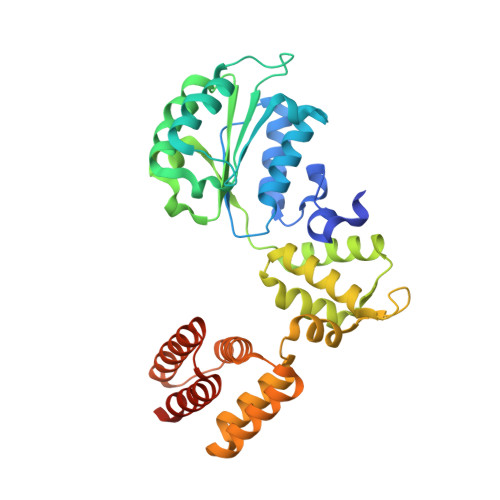
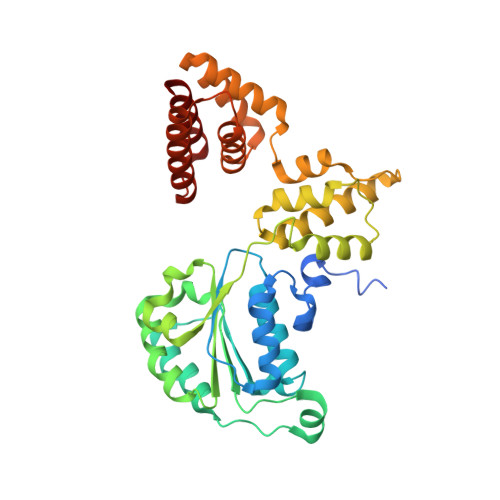
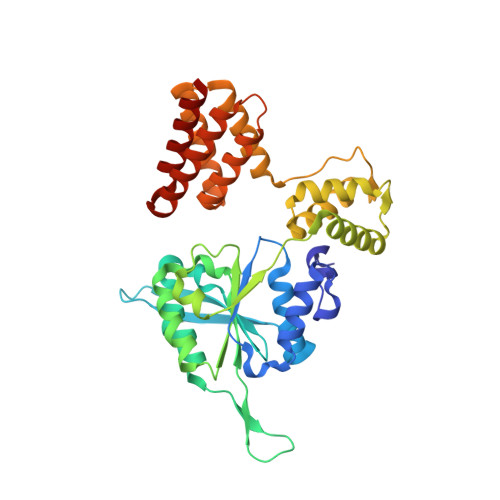
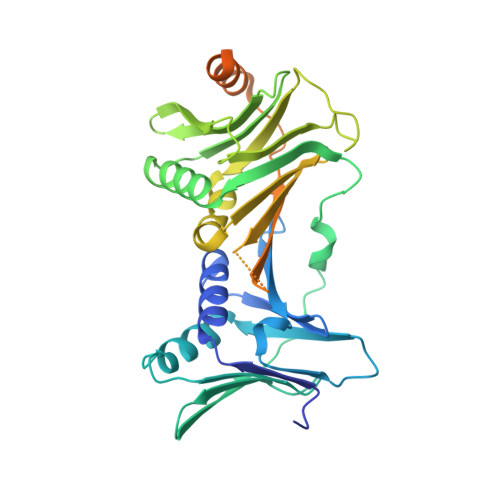
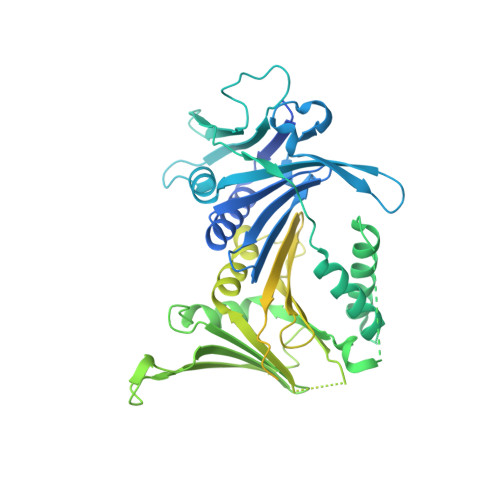
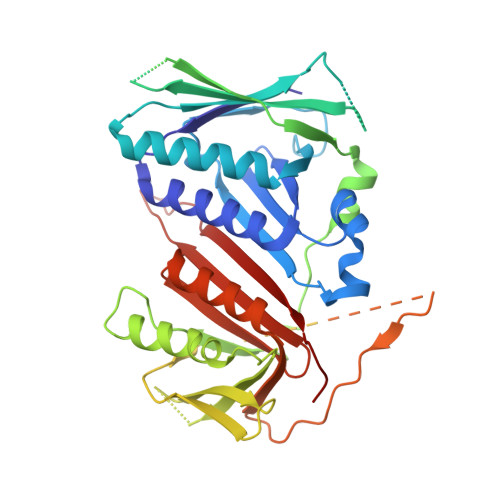
Mechanisms of loading and release of the 9-1-1 checkpoint clamp.
Castaneda, J.C., Schrecker, M., Remus, D., Hite, R.K.(2022) Nat Struct Mol Biol 29: 369-375
- PubMed: 35314831
- DOI: https://doi.org/10.1038/s41594-022-00741-7
- Primary Citation of Related Structures:
7ST9, 7STB, 7STE - PubMed Abstract:
Single-stranded or double-stranded DNA junctions with recessed 5' ends serve as loading sites for the checkpoint clamp, 9-1-1, which mediates activation of the apical checkpoint kinase, ATR Mec1 . However, the basis for 9-1-1's recruitment to 5' junctions is unclear. Here, we present structures of the yeast checkpoint clamp loader, Rad24-replication factor C (RFC), in complex with 9-1-1 and a 5' junction and in a post-ATP-hydrolysis state. Unexpectedly, 9-1-1 adopts both closed and planar open states in the presence of Rad24-RFC and DNA. Moreover, Rad24-RFC associates with the DNA junction in the opposite orientation of processivity clamp loaders with Rad24 exclusively coordinating the double-stranded region. ATP hydrolysis stimulates conformational changes in Rad24-RFC, leading to disengagement of DNA-loaded 9-1-1. Together, these structures explain 9-1-1's recruitment to 5' junctions and reveal new principles of sliding clamp loading.
- Molecular Biology Program, Memorial Sloan Kettering Cancer Center, New York, NY, USA.
Organizational Affiliation: