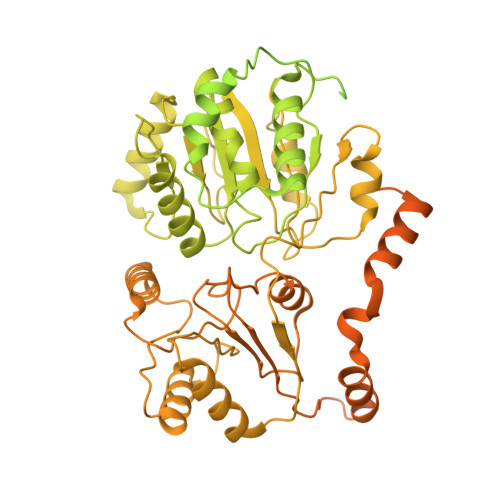
Mechanism of transcription modulation by the transcription-repair coupling factor.
Paudel, B.P., Xu, Z.Q., Jergic, S., Oakley, A.J., Sharma, N., Brown, S.H.J., Bouwer, J.C., Lewis, P.J., Dixon, N.E., van Oijen, A.M., Ghodke, H.(2022) Nucleic Acids Res 50: 5688-5712
- PubMed: 35641110
- DOI: https://doi.org/10.1093/nar/gkac449
- Primary Citation of Related Structures:
7SSG - PubMed Abstract:
Elongation by RNA polymerase is dynamically modulated by accessory factors. The transcription-repair coupling factor (TRCF) recognizes paused/stalled RNAPs and either rescues transcription or initiates transcription termination. Precisely how TRCFs choose to execute either outcome remains unclear. With Escherichia coli as a model, we used single-molecule assays to study dynamic modulation of elongation by Mfd, the bacterial TRCF. We found that nucleotide-bound Mfd converts the elongation complex (EC) into a catalytically poised state, presenting the EC with an opportunity to restart transcription. After long-lived residence in this catalytically poised state, ATP hydrolysis by Mfd remodels the EC through an irreversible process leading to loss of the RNA transcript. Further, biophysical studies revealed that the motor domain of Mfd binds and partially melts DNA containing a template strand overhang. The results explain pathway choice determining the fate of the EC and provide a molecular mechanism for transcription modulation by TRCF.
- Molecular Horizons and School of Chemistry and Molecular Bioscience, University of Wollongong, Wollongong, NSW 2522, Australia.
Organizational Affiliation: