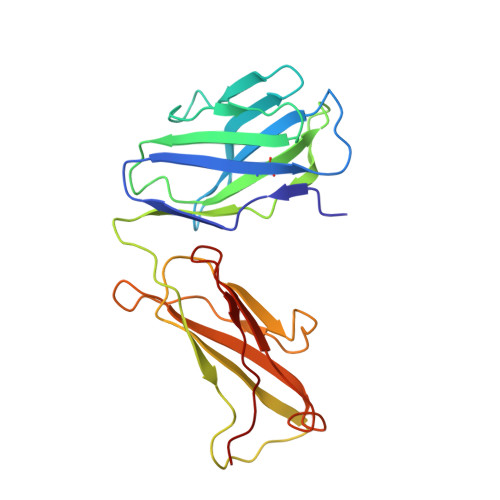
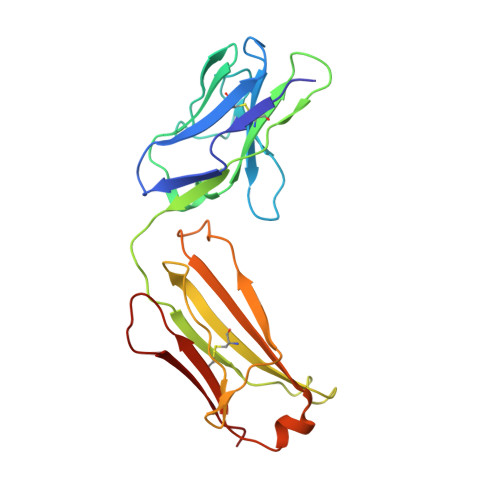
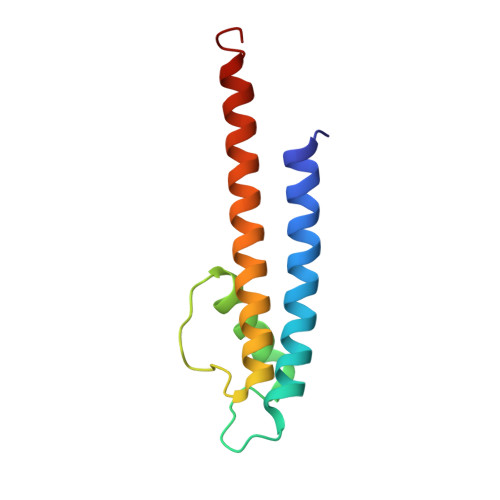
The nonconducting W434F mutant adopts upon membrane depolarization an inactivated-like state that differs from wild-type Shaker-IR potassium channels.
Coonen, L., Martinez-Morales, E., Van De Sande, D.V., Snyders, D.J., Cortes, D.M., Cuello, L.G., Labro, A.J.(2022) Sci Adv 8: eabn1731-eabn1731
- PubMed: 36112676
- DOI: https://doi.org/10.1126/sciadv.abn1731
- Primary Citation of Related Structures:
7SQW - PubMed Abstract:
Voltage-gated K + (Kv) channels mediate the flow of K + across the cell membrane by regulating the conductive state of their activation gate (AG). Several Kv channels display slow C-type inactivation, a process whereby their selectivity filter (SF) becomes less or nonconductive. It has been proposed that, in the fast inactivation-removed Shaker-IR channel, the W434F mutation epitomizes the C-type inactivated state because it functionally accelerates this process. By introducing another pore mutation that prevents AG closure, P475D, we found a way to record ionic currents of the Shaker-IR-W434F-P475D mutant at hyperpolarized membrane potentials as the W434F-mutant SF recovers from its inactivated state. This W434F conductive state lost its high K + over Na + selectivity, and even NMDG + can permeate, features not observed in a wild-type SF. This indicates that, at least during recovery from inactivation, the W434F-mutant SF transitions to a widened and noncationic specific conformation.
- Department of Biomedical Sciences, Faculty of Pharmaceutical, Biomedical and Veterinary Sciences, University of Antwerp, 2610 Antwerp, Belgium.
Organizational Affiliation: