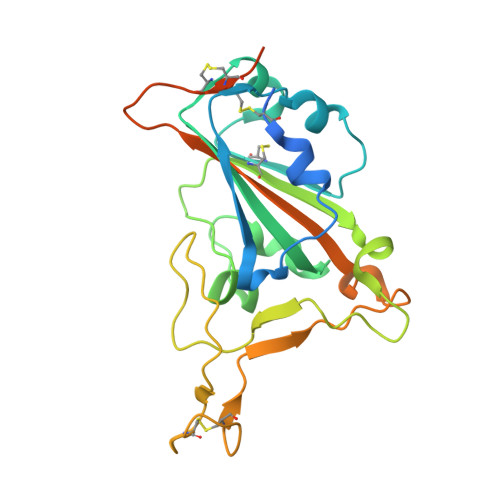
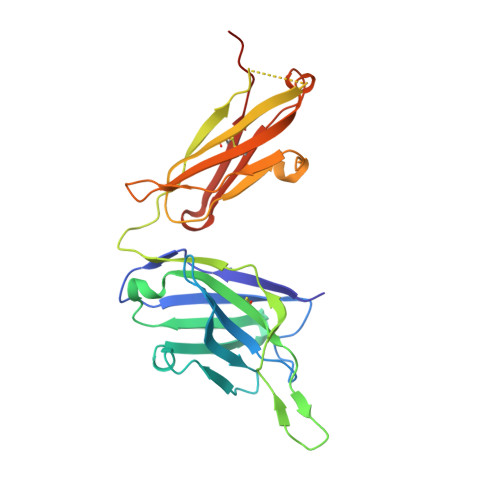
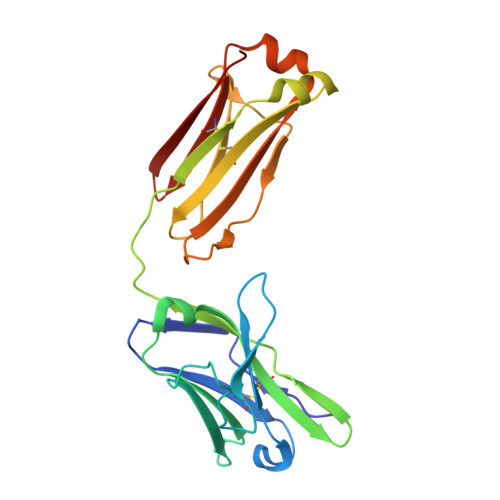
An antibody class with a common CDRH3 motif broadly neutralizes sarbecoviruses.
Liu, L., Iketani, S., Guo, Y., Reddem, E.R., Casner, R.G., Nair, M.S., Yu, J., Chan, J.F., Wang, M., Cerutti, G., Li, Z., Morano, N.C., Castagna, C.D., Corredor, L., Chu, H., Yuan, S., Poon, V.K., Chan, C.C., Chen, Z., Luo, Y., Cunningham, M., Chavez, A., Yin, M.T., Perlin, D.S., Tsuji, M., Yuen, K.Y., Kwong, P.D., Sheng, Z., Huang, Y., Shapiro, L., Ho, D.D.(2022) Sci Transl Med 14: eabn6859-eabn6859
- PubMed: 35438546
- DOI: https://doi.org/10.1126/scitranslmed.abn6859
- Primary Citation of Related Structures:
7SD5, 7SI2, 7TTM, 7TTX, 7TTY - PubMed Abstract:
The devastation caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has made clear the importance of pandemic preparedness. To address future zoonotic outbreaks due to related viruses in the sarbecovirus subgenus, we identified a human monoclonal antibody, 10-40, that neutralized or bound all sarbecoviruses tested in vitro and protected against SARS-CoV-2 and SARS-CoV in vivo. Comparative studies with other receptor-binding domain (RBD)-directed antibodies showed 10-40 to have the greatest breadth against sarbecoviruses, suggesting that 10-40 is a promising agent for pandemic preparedness. Moreover, structural analyses on 10-40 and similar antibodies not only defined an epitope cluster in the inner face of the RBD that is well conserved among sarbecoviruses but also uncovered a distinct antibody class with a common CDRH3 motif. Our analyses also suggested that elicitation of this class of antibodies may not be overly difficult, an observation that bodes well for the development of a pan-sarbecovirus vaccine.
- Aaron Diamond AIDS Research Center, Columbia University Vagelos College of Physicians and Surgeons, New York, NY 10032, USA.
Organizational Affiliation: