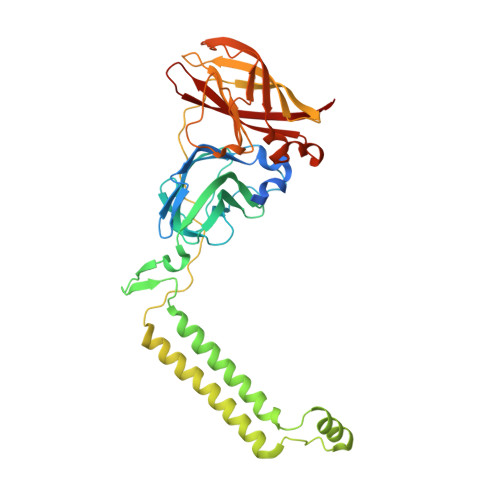
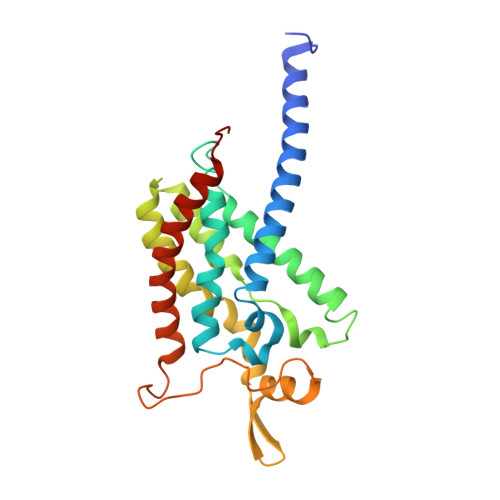
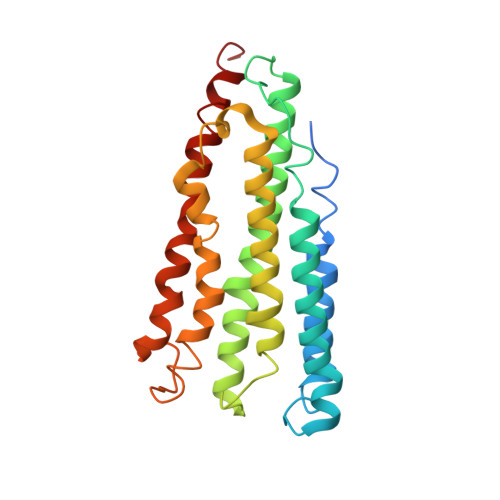
Recovery of particulate methane monooxygenase structure and activity in a lipid bilayer.
Koo, C.W., Tucci, F.J., He, Y., Rosenzweig, A.C.(2022) Science 375: 1287-1291
- PubMed: 35298269
- DOI: https://doi.org/10.1126/science.abm3282
- Primary Citation of Related Structures:
7S4H, 7S4I, 7S4J, 7S4K, 7S4L, 7S4M, 7T4O, 7T4P - PubMed Abstract:
Bacterial methane oxidation using the enzyme particulate methane monooxygenase (pMMO) contributes to the removal of environmental methane, a potent greenhouse gas. Crystal structures determined using inactive, detergent-solubilized pMMO lack several conserved regions neighboring the proposed active site. We show that reconstituting pMMO in nanodiscs with lipids extracted from the native organism restores methane oxidation activity. Multiple nanodisc-embedded pMMO structures determined by cryo-electron microscopy to 2.14- to 2.46-angstrom resolution reveal the structure of pMMO in a lipid environment. The resulting model includes stabilizing lipids, regions of the PmoA and PmoC subunits not observed in prior structures, and a previously undetected copper-binding site in the PmoC subunit with an adjacent hydrophobic cavity. These structures provide a revised framework for understanding and engineering pMMO function.
- Department of Molecular Biosciences, Northwestern University, Evanston, IL 60208, USA.
Organizational Affiliation: