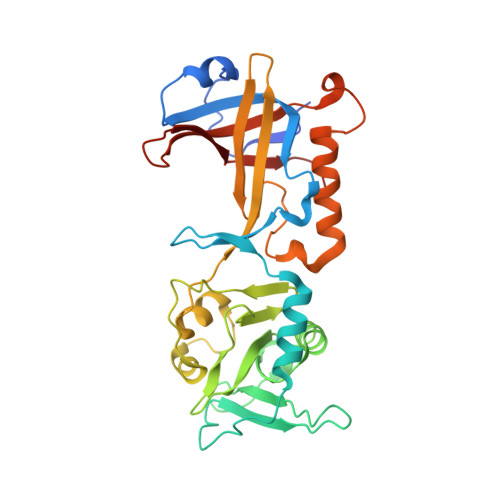
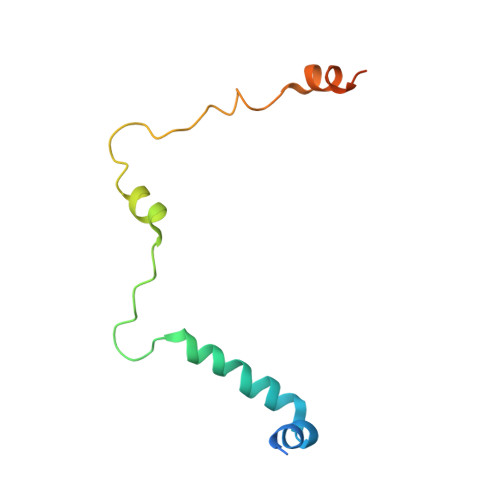
POT1-TPP1 binding stabilizes POT1, promoting efficient telomere maintenance.
Aramburu, T., Kelich, J., Rice, C., Skordalakes, E.(2022) Comput Struct Biotechnol J 20: 675-684
- PubMed: 35140887
- DOI: https://doi.org/10.1016/j.csbj.2022.01.005
- Primary Citation of Related Structures:
7S1O, 7S1T, 7S1U - PubMed Abstract:
Telomeric POT1-TPP1 binding is critical to telomere maintenance and disruption of this complex may lead to cancer. Current data suggests a reduction of intracellular POT1 levels in the absence of TPP1. Here we provide evidence of POT1 plasticity that contributes to its lack of stability in the absence of TPP1 binding. Structural data reveals inter- and intramolecular POT1C domain flexibility in the absence of TPP1. Thermostability and proteolytic resistance assays show that POT1C and the mutant complex POT1C(Q623H)-TPP1(PBD) are less stable than the wild type POT1C-TPP1(PBD), suggesting that TPP1 binding to POT1 stabilizes POT1C and makes it less accessible to proteasomal degradation in the cell. Disruption of the POT1-TPP1 complex such as through cancer-associated mutations leads to a reduction of intracellular POT1, telomere uncapping, and telomere associated DNA damage response (DDR). DDR in turn leads to senescence or genomic instability and oncogenesis.
- The Wistar Institute, 3601 Spruce St, Philadelphia, PA 19104, USA.
Organizational Affiliation: