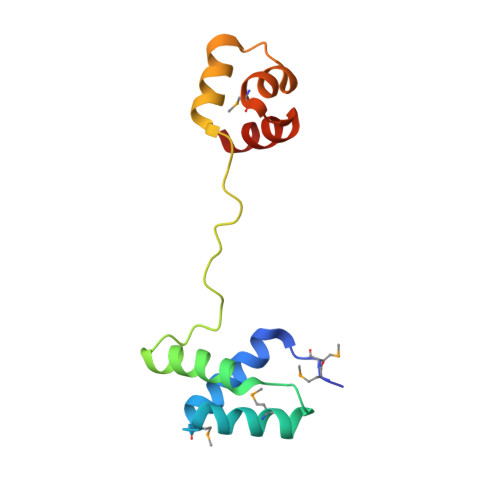
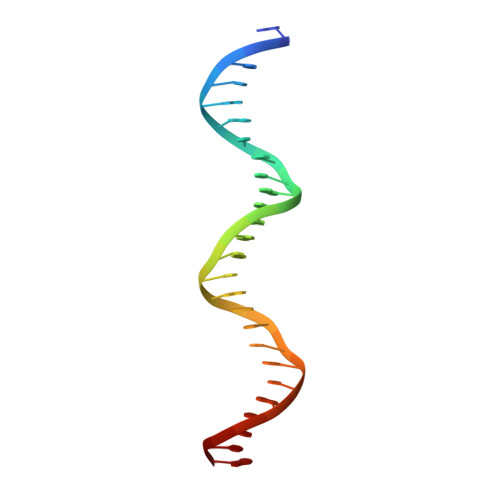
Structural and genome-wide analyses suggest that transposon-derived protein SETMAR alters transcription and splicing.
Chen, Q., Bates, A.M., Hanquier, J.N., Simpson, E., Rusch, D.B., Podicheti, R., Liu, Y., Wek, R.C., Cornett, E.M., Georgiadis, M.M.(2022) J Biological Chem 298: 101894-101894
- PubMed: 35378129
- DOI: https://doi.org/10.1016/j.jbc.2022.101894
- Primary Citation of Related Structures:
7S03 - PubMed Abstract:
Extensive portions of the human genome have unknown function, including those derived from transposable elements. One such element, the DNA transposon Hsmar1, entered the primate lineage approximately 50 million years ago leaving behind terminal inverted repeat (TIR) sequences and a single intact copy of the Hsmar1 transposase, which retains its ancestral TIR-DNA-binding activity, and is fused with a lysine methyltransferase SET domain to constitute the chimeric SETMAR gene. Here, we provide a structural basis for recognition of TIRs by SETMAR and investigate the function of SETMAR through genome-wide approaches. As elucidated in our 2.37 Å crystal structure, SETMAR forms a dimeric complex with each DNA-binding domain bound specifically to TIR-DNA through the formation of 32 hydrogen bonds. We found that SETMAR recognizes primarily TIR sequences (∼5000 sites) within the human genome as assessed by chromatin immunoprecipitation sequencing analysis. In two SETMAR KO cell lines, we identified 163 shared differentially expressed genes and 233 shared alternative splicing events. Among these genes are several pre-mRNA-splicing factors, transcription factors, and genes associated with neuronal function, and one alternatively spliced primate-specific gene, TMEM14B, which has been identified as a marker for neocortex expansion associated with brain evolution. Taken together, our results suggest a model in which SETMAR impacts differential expression and alternative splicing of genes associated with transcription and neuronal function, potentially through both its TIR-specific DNA-binding and lysine methyltransferase activities, consistent with a role for SETMAR in simian primate development.
- Department of Biochemistry and Molecular Biology, Indiana University School of Medicine, Indianapolis, Indiana, USA.
Organizational Affiliation: