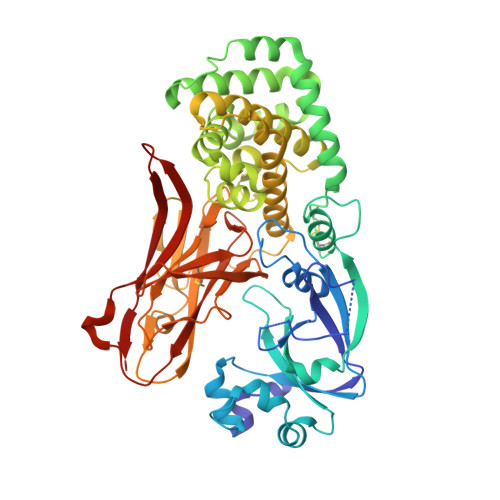
Monomeric crystal structure of the vaccine carrier protein CRM 197 and implications for vaccine development.
Gallagher, D.T., Oganesyan, N., Lees, A.(2023) Acta Crystallogr F Struct Biol Commun 79: 82-86
- PubMed: 36995122
- DOI: https://doi.org/10.1107/S2053230X23002364
- Primary Citation of Related Structures:
7RRW - PubMed Abstract:
CRM 197 is a genetically detoxified mutant of diphtheria toxin (DT) that is widely used as a carrier protein in conjugate vaccines. Protective immune responses to several bacterial diseases are obtained by coupling CRM 197 to glycans from these pathogens. Wild-type DT has been described in two oligomeric forms: a monomer and a domain-swapped dimer. Their proportions depend on the chemical conditions and especially the pH, with a large kinetic barrier to interconversion. A similar situation occurs in CRM 197 , where the monomer is preferred for vaccine synthesis. Despite 30 years of research and the increasing application of CRM 197 in conjugate vaccines, until now all of its available crystal structures have been dimeric. Here, CRM 197 was expressed as a soluble, intracellular protein in an Escherichia coli strain engineered to have an oxidative cytoplasm. The purified product, called EcoCRM, remained monomeric throughout crystallization. The structure of monomeric EcoCRM is reported at 2.0 Å resolution with the domain-swapping hinge loop (residues 379-387) in an extended, exposed conformation, similar to monomeric wild-type DT. The structure enables comparisons across expression systems and across oligomeric states, with implications for monomer-dimer interconversion and for the optimization of conjugation.
- National Institute of Standards and Technology, 9600 Gudelsky Drive, Rockville, MD 20850, USA.
Organizational Affiliation: