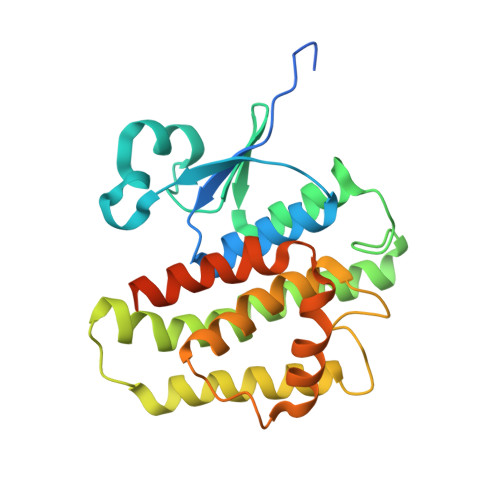
Architecture and potential roles of a delta-class glutathione S-transferase in protecting honey bee from agrochemicals.
Moural, T.W., Koirala B K, S., Bhattarai, G., He, Z., Guo, H., Phan, N.T., Rajotte, E.G., Biddinger, D.J., Hoover, K., Zhu, F.(2023) Chemosphere 350: 141089-141089
- PubMed: 38163465
- DOI: https://doi.org/10.1016/j.chemosphere.2023.141089
- Primary Citation of Related Structures:
7RHP - PubMed Abstract:
The European honey bee, Apis mellifera, serves as the principle managed pollinator species globally. In recent decades, honey bee populations have been facing serious health threats from combined biotic and abiotic stressors, including diseases, limited nutrition, and agrochemical exposure. Understanding the molecular mechanisms underlying xenobiotic adaptation of A. mellifera is critical, considering its extensive exposure to phytochemicals and agrochemicals present in the environment. In this study, we conducted a comprehensive structural and functional characterization of AmGSTD1, a delta class glutathione S-transferase (GST), to unravel its roles in agrochemical detoxification and antioxidative stress responses. We determined the 3-dimensional (3D) structure of a honey bee GST using protein crystallography for the first time, providing new insights into its molecular structure. Our investigations revealed that AmGSTD1 metabolizes model substrates, including 1-chloro-2,4-dinitrobenzene (CDNB), p-nitrophenyl acetate (PNA), phenylethyl isothiocyanate (PEITC), propyl isothiocyanate (PITC), and the oxidation byproduct 4-hydroxynonenal (HNE). Moreover, we discovered that AmGSTD1 exhibits binding affinity with the fluorophore 8-Anilinonaphthalene-1-sulfonic acid (ANS), which can be inhibited with various herbicides, fungicides, insecticides, and their metabolites. These findings highlight the potential contribution of AmGSTD1 in safeguarding honey bee health against various agrochemicals, while also mitigating oxidative stress resulting from exposure to these substances.
- Department of Entomology, Pennsylvania State University, University Park, PA 16802, USA. Electronic address: twm78@psu.edu.
Organizational Affiliation: