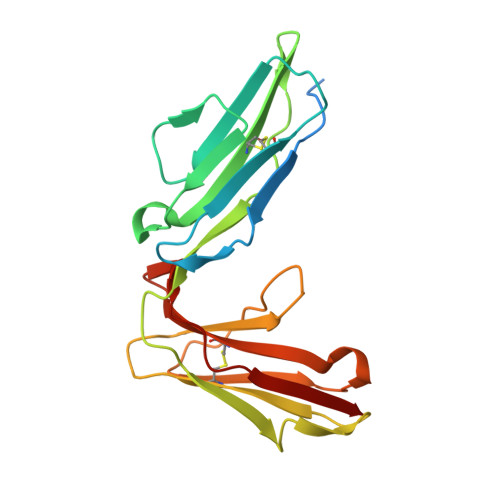
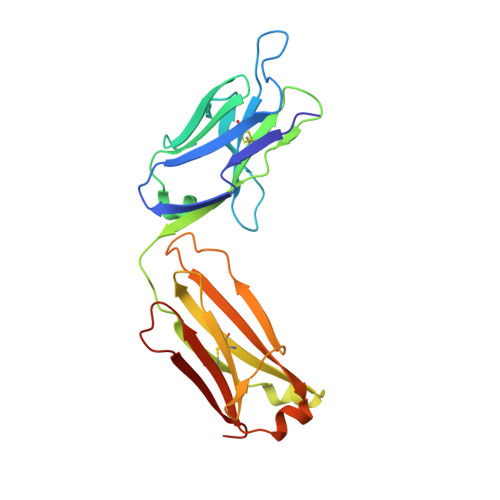
Targeting platelet GPVI with glenzocimab: a novel mechanism for inhibition.
Billiald, P., Slater, A., Welin, M., Clark, J.C., Loyau, S., Pugniere, M., Jiacomini, I.G., Rose, N., Lebozec, K., Toledano, E., Francois, D., Watson, S.P., Jandrot-Perrus, M.(2023) Blood Adv 7: 1258-1268
- PubMed: 36375047
- DOI: https://doi.org/10.1182/bloodadvances.2022007863
- Primary Citation of Related Structures:
7R58 - PubMed Abstract:
Platelet glycoprotein VI (GPVI) is attracting interest as a potential target for the development of new antiplatelet molecules with a low bleeding risk. GPVI binding to vascular collagen initiates thrombus formation and GPVI interactions with fibrin promote the growth and stability of the thrombus. In this study, we show that glenzocimab, a clinical stage humanized antibody fragment (Fab) with a high affinity for GPVI, blocks the binding of both ligands through a combination of steric hindrance and structural change. A cocrystal of glenzocimab with an extracellular domain of monomeric GPVI was obtained and its structure determined to a resolution of 1.9 Å. The data revealed that (1) glenzocimab binds to the D2 domain of GPVI, GPVI dimerization was not observed in the crystal structure because glenzocimab prevented D2 homotypic interactions and the formation of dimers that have a high affinity for collagen and fibrin; and (2) the light variable domain of the GPVI-bound Fab causes steric hindrance that is predicted to prevent the collagen-related peptide (CRP)/collagen fibers from extending out of their binding site and preclude GPVI clustering and downstream signaling. Glenzocimab did not bind to a truncated GPVI missing loop residues 129 to 136, thus validating the epitope identified in the crystal structure. Overall, these findings demonstrate that the binding of glenzocimab to the D2 domain of GPVI induces steric hindrance and structural modifications that drive the inhibition of GPVI interactions with its major ligands.
- Laboratory for Vascular Translational Science, UMR_S1148 INSERM, Université Paris Cité, Hôpital Bichat, Paris, France.
Organizational Affiliation: