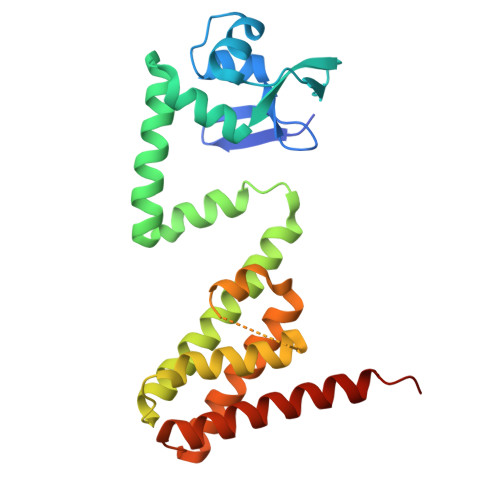
Structural and functional characterization of DdrC, a novel DNA damage-induced nucleoid associated protein involved in DNA compaction.
Banneville, A.S., Bouthier de la Tour, C., De Bonis, S., Hognon, C., Colletier, J.P., Teulon, J.M., Le Roy, A., Pellequer, J.L., Monari, A., Dehez, F., Confalonieri, F., Servant, P., Timmins, J.(2022) Nucleic Acids Res 50: 7680-7696
- PubMed: 35801857
- DOI: https://doi.org/10.1093/nar/gkac563
- Primary Citation of Related Structures:
7QVB - PubMed Abstract:
Deinococcus radiodurans is a spherical bacterium well-known for its outstanding resistance to DNA-damaging agents. Exposure to such agents leads to drastic changes in the transcriptome of D. radiodurans. In particular, four Deinococcus-specific genes, known as DNA Damage Response genes, are strongly up-regulated and have been shown to contribute to the resistance phenotype of D. radiodurans. One of these, DdrC, is expressed shortly after exposure to γ-radiation and is rapidly recruited to the nucleoid. In vitro, DdrC has been shown to compact circular DNA, circularize linear DNA, anneal complementary DNA strands and protect DNA from nucleases. To shed light on the possible functions of DdrC in D. radiodurans, we determined the crystal structure of the domain-swapped DdrC dimer at a resolution of 2.5 Å and further characterized its DNA binding and compaction properties. Notably, we show that DdrC bears two asymmetric DNA binding sites located on either side of the dimer and can modulate the topology and level of compaction of circular DNA. These findings suggest that DdrC may be a DNA damage-induced nucleoid-associated protein that enhances nucleoid compaction to limit the dispersion of the fragmented genome and facilitate DNA repair after exposure to severe DNA damaging conditions.
- Univ. Grenoble Alpes, CEA, CNRS, IBS, F-38000 Grenoble, France.
Organizational Affiliation: