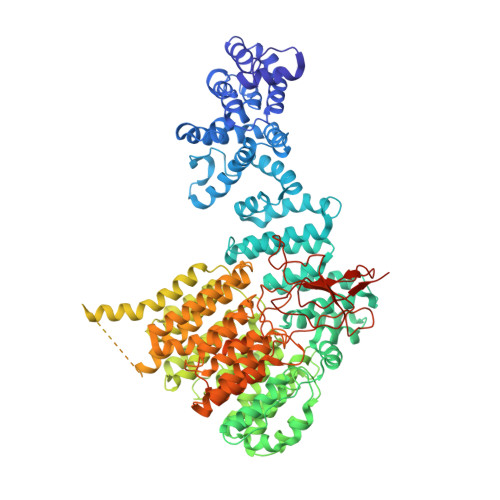
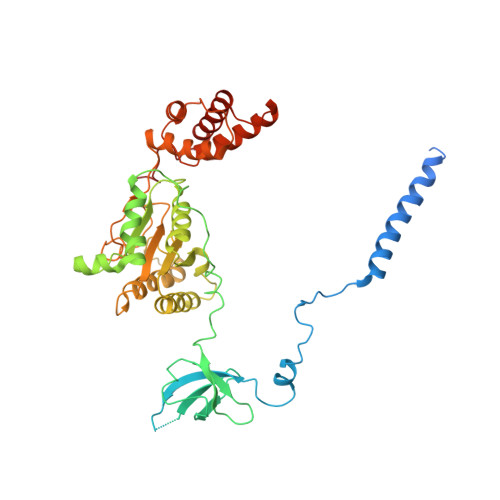
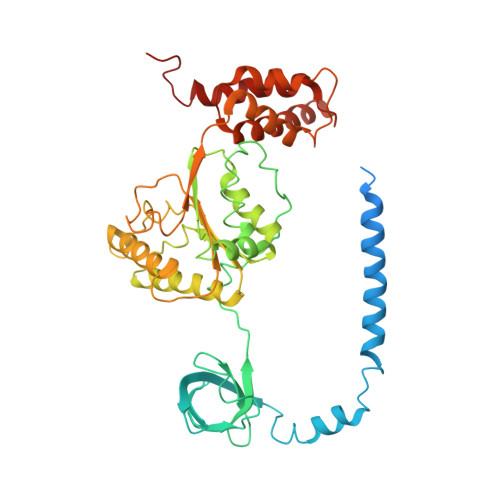
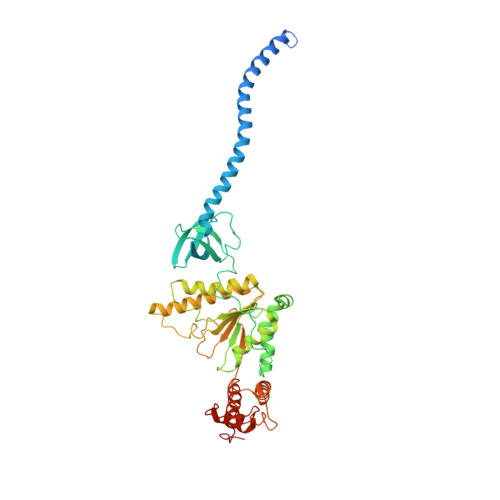
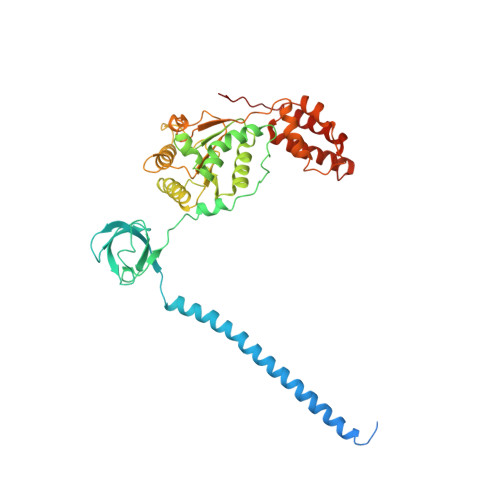
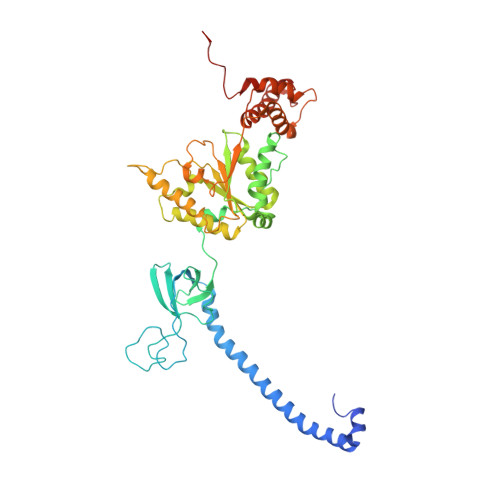
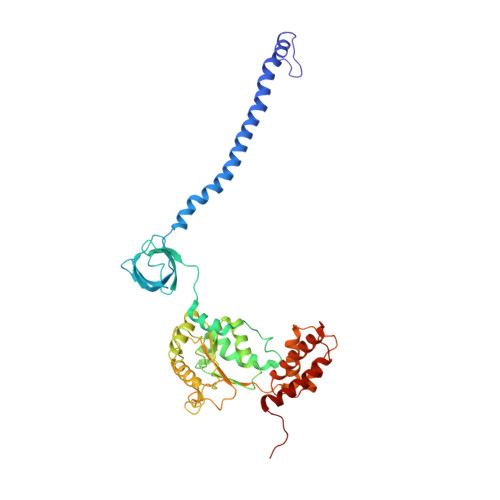
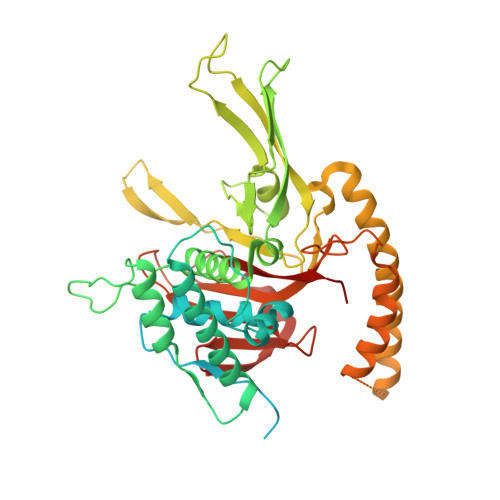
Allosteric control of Ubp6 and the proteasome via a bidirectional switch.
Hung, K.Y.S., Klumpe, S., Eisele, M.R., Elsasser, S., Tian, G., Sun, S., Moroco, J.A., Cheng, T.C., Joshi, T., Seibel, T., Van Dalen, D., Feng, X.H., Lu, Y., Ovaa, H., Engen, J.R., Lee, B.H., Rudack, T., Sakata, E., Finley, D.(2022) Nat Commun 13: 838